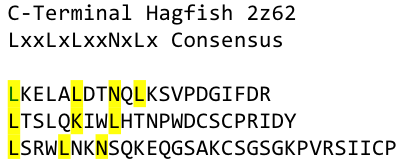Team:Freiburg/Modelling
From 2011.igem.org
(Difference between revisions)
(→Modelling: Rational protein design) |
|||
| Line 31: | Line 31: | ||
|} | |} | ||
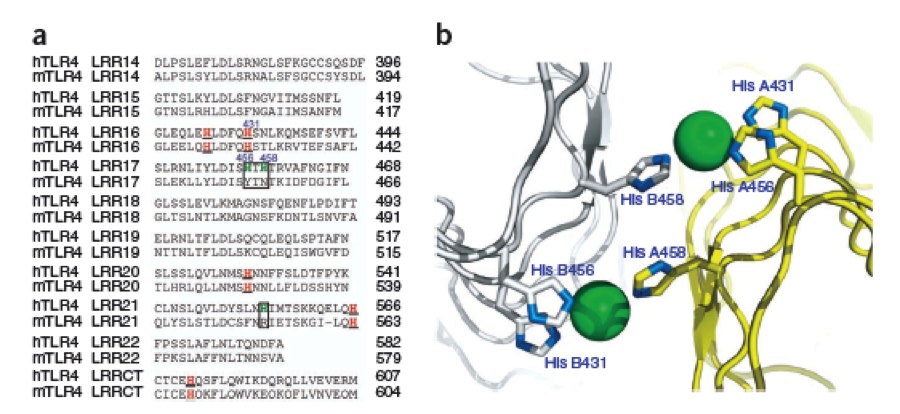
By analyzing the logo it was obvious which positions of the LRR motif were conserved and which not – AND: which of the non conserved aminoacids appeared in what kind of patterns. Were there positions in the protein that required a polar aminoacid? Or non polar, hydrophobic /-philic, charged, non-charged?. We compared the consensus sequence with the 3D structure, using PYMOL, to extract as much information as possible and then came up with this ideal consensus sequence: | By analyzing the logo it was obvious which positions of the LRR motif were conserved and which not – AND: which of the non conserved aminoacids appeared in what kind of patterns. Were there positions in the protein that required a polar aminoacid? Or non polar, hydrophobic /-philic, charged, non-charged?. We compared the consensus sequence with the 3D structure, using PYMOL, to extract as much information as possible and then came up with this ideal consensus sequence: | ||
| - | [[File:Freiburg11_Seq2.png | + | {| style="color:black; background-color:lightgrey;" cellpadding="10%" cellpadding="15%" cellspacing="0" border="1" |
| + | |[[File:Freiburg11_Seq2.png|300px]] | ||
| + | Caption | ||
| + | |} | ||
The next consideration we had to do, was: how many Nickel do we need on the surface of our ideal Nickel binding protein, in what pattern, with what distances between, and at what angles towards each other to allow proper ion complexation? | The next consideration we had to do, was: how many Nickel do we need on the surface of our ideal Nickel binding protein, in what pattern, with what distances between, and at what angles towards each other to allow proper ion complexation? | ||
| Line 40: | Line 43: | ||
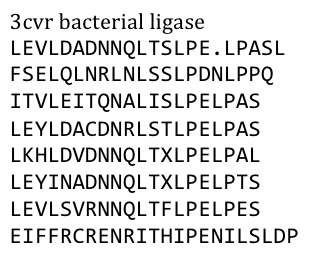
To get a first impression how Histidines are positioned in a bacterial Ligase, we replaced all non conserved aminoacids by Histidines and sent the sequence for a 3D sequence prediction to I-TASSER, an online structure prediction software tool. | To get a first impression how Histidines are positioned in a bacterial Ligase, we replaced all non conserved aminoacids by Histidines and sent the sequence for a 3D sequence prediction to I-TASSER, an online structure prediction software tool. | ||
| - | + | {| style="color:black; background-color:lightgrey;" cellpadding="10%" cellpadding="15%" cellspacing="0" border="1" | |
| - | [[File:Freiburg11_Seq7.png | + | |[[File:Freiburg11_Seq7.png|300px]] |
| - | + | Caption | |
| + | |} | ||
| Line 58: | Line 62: | ||
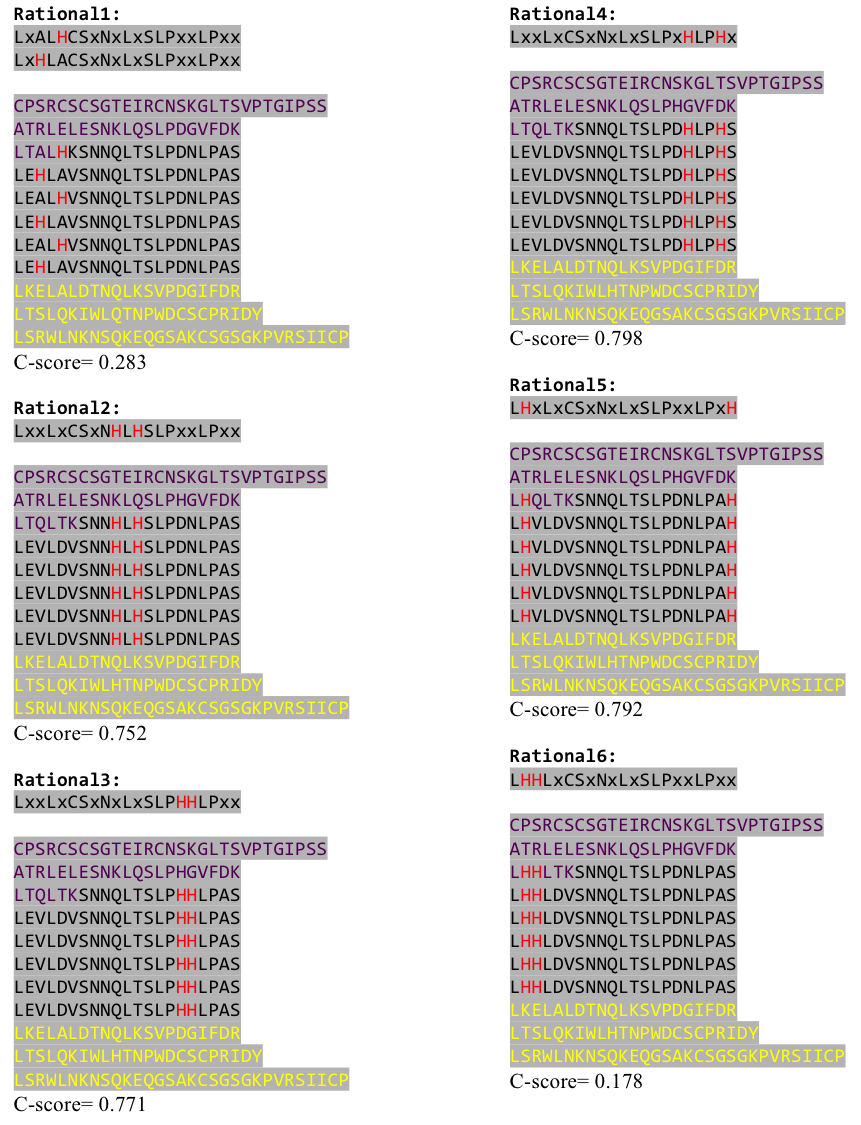
The predicted models are all evaluated by the C-Score, which assesses the quality of the prediction. It has a value from -5 to 2; the more positive the C-Score the merrier and more plausible the predicted structure is. | The predicted models are all evaluated by the C-Score, which assesses the quality of the prediction. It has a value from -5 to 2; the more positive the C-Score the merrier and more plausible the predicted structure is. | ||
| - | [[File:Freiburg11Modelling3.png | + | [[File:Freiburg11Modelling3.png|750px]] |
| + | Caption | ||
1.Ambrish Roy, Alper Kucukural, Yang Zhang. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, vol 5, 725-738 (2010). | 1.Ambrish Roy, Alper Kucukural, Yang Zhang. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, vol 5, 725-738 (2010). | ||
| Line 70: | Line 75: | ||
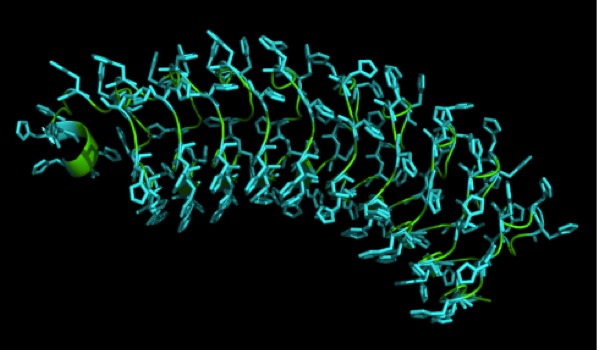
We received this pdb file with a C-score= 0.53. Obviously this was not a realistic model, since the folding would surely be impaired by so many Histidines everywhere. But is was useful to find several good locations, were the Nickel could fit in in respect to the requirements it has for binding ligands. | We received this pdb file with a C-score= 0.53. Obviously this was not a realistic model, since the folding would surely be impaired by so many Histidines everywhere. But is was useful to find several good locations, were the Nickel could fit in in respect to the requirements it has for binding ligands. | ||
| - | [[File:Freiburg11Modelling4.png | + | {| style="color:black; background-color:lightgrey;" cellpadding="10%" cellpadding="15%" cellspacing="0" border="1" |
| - | + | |[[File:Freiburg11Modelling4.png|400px]] | |
| + | Caption | ||
| + | |} | ||
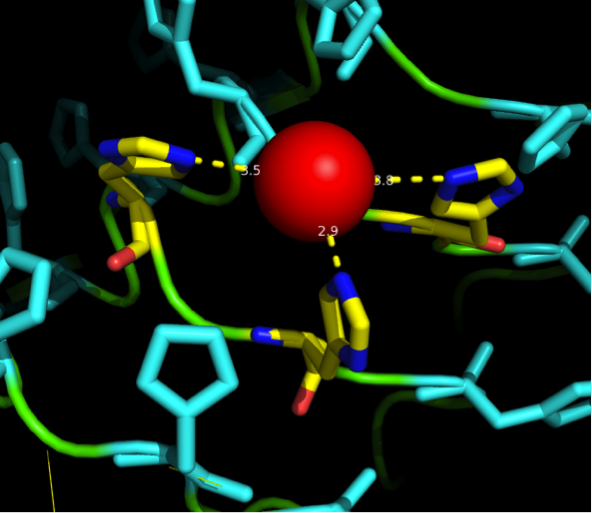
We manually fit in a Nickel ion, as it was crystallized in the mentioned PDB file, and measured the distances and evaluated the 3dimensional orientation of the Histidines towards the Nickel. Histidines can coordinate ligands with their free electron pair pointing planar away from the imidazole ring. | We manually fit in a Nickel ion, as it was crystallized in the mentioned PDB file, and measured the distances and evaluated the 3dimensional orientation of the Histidines towards the Nickel. Histidines can coordinate ligands with their free electron pair pointing planar away from the imidazole ring. | ||
| - | [[File:Freiburg11Modelling5.png | + | {| style="color:black; background-color:lightgrey;" cellpadding="10%" cellpadding="15%" cellspacing="0" border="1" |
| - | + | |[[File:Freiburg11Modelling5.png|400px]] | |
| + | Caption | ||
| + | |} | ||
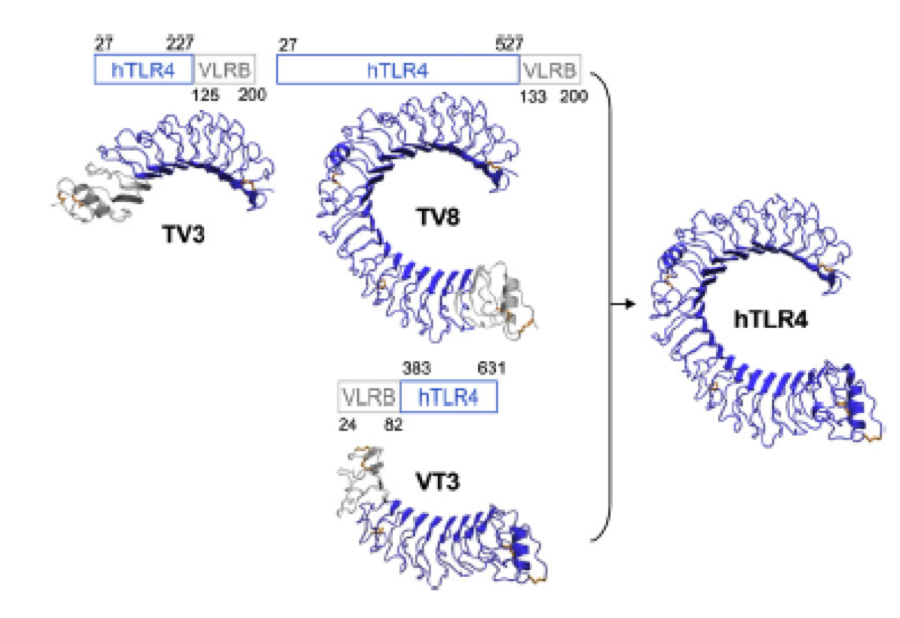
What we further realized from the structure file was, that the end of the LRR segments were “open”, that means, the hydrophobic core of the protein was exposed and as it is visible in the prediction, curled in on one end into a sort of helix. This means the protein folding is not reliable and the structure needs some caps on both ends to stabilize the LRR core motif. A solution to this was shown by Schmidt et al 2010, who crystallized the TLR-4 receptor. | What we further realized from the structure file was, that the end of the LRR segments were “open”, that means, the hydrophobic core of the protein was exposed and as it is visible in the prediction, curled in on one end into a sort of helix. This means the protein folding is not reliable and the structure needs some caps on both ends to stabilize the LRR core motif. A solution to this was shown by Schmidt et al 2010, who crystallized the TLR-4 receptor. | ||
Revision as of 16:36, 20 September 2011
 "
"
 Contact
Contact