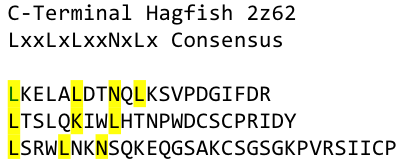Team:Freiburg/Modelling
From 2011.igem.org
(Difference between revisions)
(→Modelling: Rational protein design) |
(→Modelling: Rational protein design) |
||
| Line 82: | Line 82: | ||
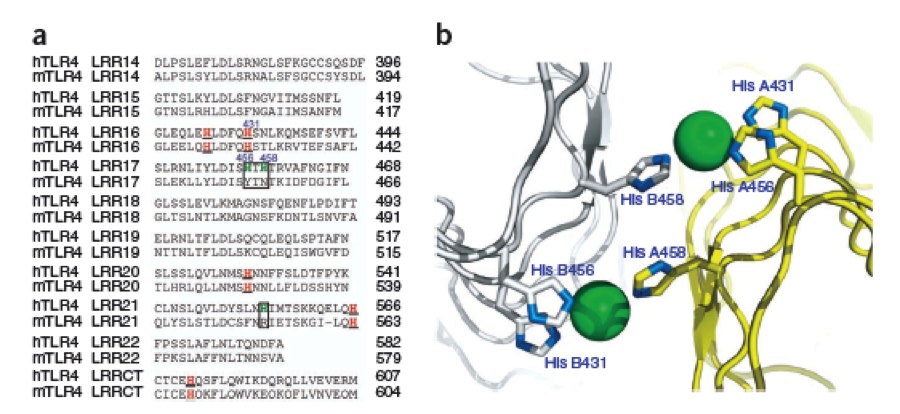
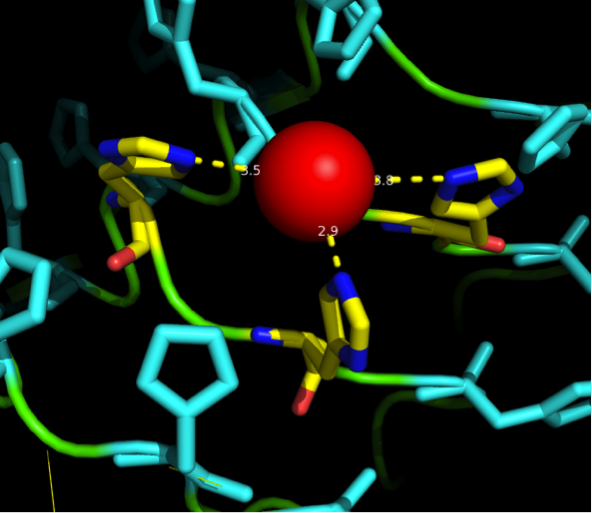
We manually fit in a Nickel ion, as it was crystallized in the mentioned PDB file, and measured the distances and evaluated the 3dimensional orientation of the Histidines towards the Nickel. Histidines can coordinate ligands with their free electron pair pointing planar away from the imidazole ring. | We manually fit in a Nickel ion, as it was crystallized in the mentioned PDB file, and measured the distances and evaluated the 3dimensional orientation of the Histidines towards the Nickel. Histidines can coordinate ligands with their free electron pair pointing planar away from the imidazole ring. | ||
| - | {| style="color:black; background-color:lightgrey;" cellpadding="10%" cellpadding="15%" cellspacing="0" border="1" align=" | + | {| style="color:black; background-color:lightgrey;" cellpadding="10%" cellpadding="15%" cellspacing="0" border="1" align="right" |
| - | |[[File:Freiburg11Modelling5.png| | + | |[[File:Freiburg11Modelling5.png|250px]] |
Caption | Caption | ||
|} | |} | ||
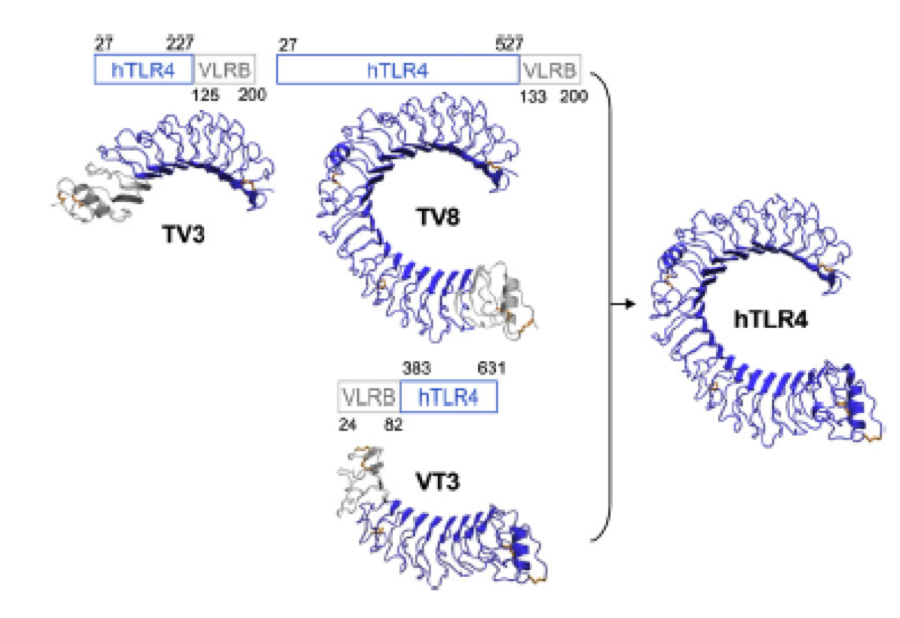
What we further realized from the structure file was, that the end of the LRR segments were “open”, that means, the hydrophobic core of the protein was exposed and as it is visible in the prediction, curled in on one end into a sort of helix. This means the protein folding is not reliable and the structure needs some caps on both ends to stabilize the LRR core motif. A solution to this was shown by Schmidt et al 2010, who crystallized the TLR-4 receptor. | What we further realized from the structure file was, that the end of the LRR segments were “open”, that means, the hydrophobic core of the protein was exposed and as it is visible in the prediction, curled in on one end into a sort of helix. This means the protein folding is not reliable and the structure needs some caps on both ends to stabilize the LRR core motif. A solution to this was shown by Schmidt et al 2010, who crystallized the TLR-4 receptor. | ||
| - | [[File:Freiburg11Modelling6.png| | + | {| style="color:black; background-color:lightgrey;" cellpadding="10%" cellpadding="15%" cellspacing="0" border="1" |
| + | |[[File:Freiburg11Modelling6.png|700px]] | ||
| + | Caption | ||
| + | |} | ||
| + | |||
In this very nice piece of work he dissected the TLR4(PDB: 3FXI) into 3 parts, since it was to large and unhandy to be crystallized at once. To overcome this problem of an exposed hydrophobic core, he used the N- and C-terminal protein fragments of a LRR protein derived from hagfish. They tried a variety of different versions of how to glue together his fragments and these N- and C-terminal caps until he found a working one. | In this very nice piece of work he dissected the TLR4(PDB: 3FXI) into 3 parts, since it was to large and unhandy to be crystallized at once. To overcome this problem of an exposed hydrophobic core, he used the N- and C-terminal protein fragments of a LRR protein derived from hagfish. They tried a variety of different versions of how to glue together his fragments and these N- and C-terminal caps until he found a working one. | ||
We used this knowledge for our purpose and took the same sequences and attached them to our protein sequence. These caps partially still show the typical LRR consensus sequences (which luckily is highly conserved in all kingdoms of life!), which made it possible to fit them onto our stack of LRR loops in the right position. | We used this knowledge for our purpose and took the same sequences and attached them to our protein sequence. These caps partially still show the typical LRR consensus sequences (which luckily is highly conserved in all kingdoms of life!), which made it possible to fit them onto our stack of LRR loops in the right position. | ||
| - | [[File:Freiburg11_Seq3.png | + | {| style="color:black; background-color:lightgrey;" cellpadding="10%" cellpadding="15%" cellspacing="0" border="1" align="right" |
| - | + | |[[File:Freiburg11_Seq3.png|300px]] | |
| + | Caption | ||
| + | |} | ||
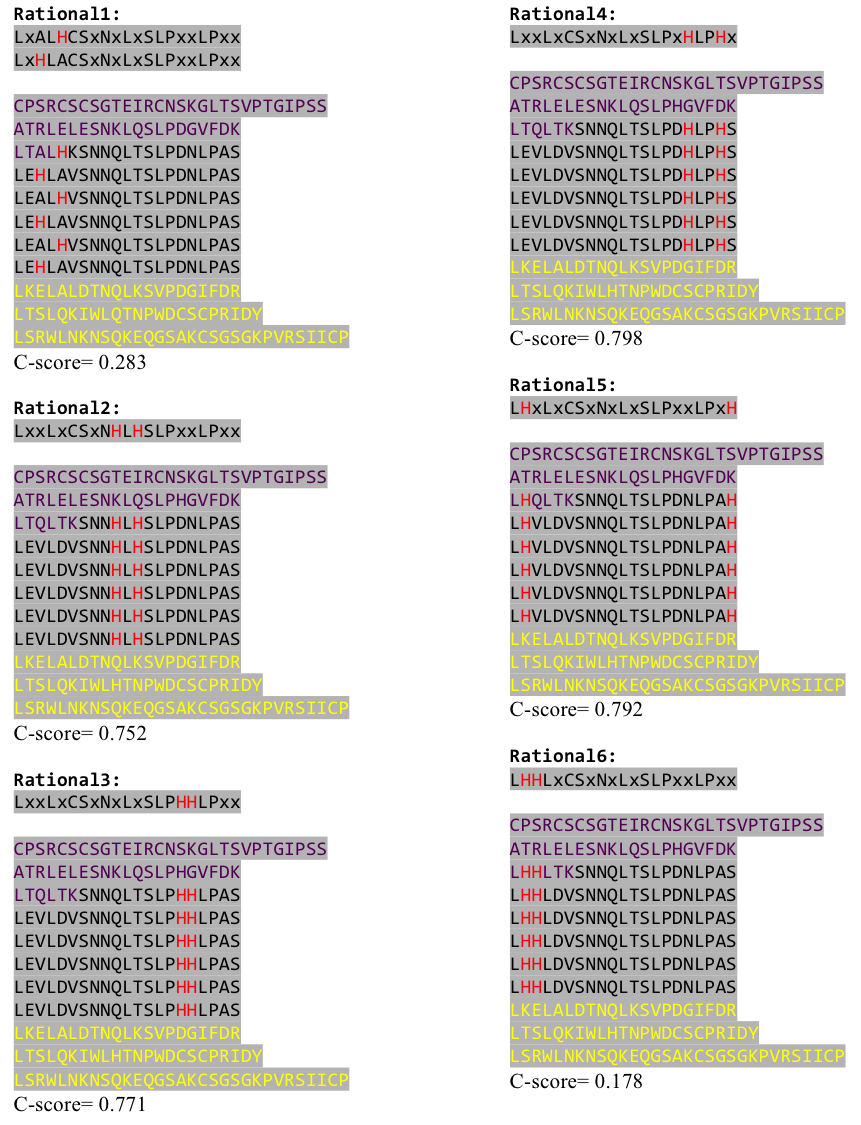
To the outer end the protein has a helix on the N-terminal side shielding off the core and the C-terminus the LRR disappears slowly turn by turn with more and more hydrophilic aminoacids replacing the LRR consensus sequence. | To the outer end the protein has a helix on the N-terminal side shielding off the core and the C-terminus the LRR disappears slowly turn by turn with more and more hydrophilic aminoacids replacing the LRR consensus sequence. | ||
| Line 100: | Line 106: | ||
{|style="color:black; border="1" width="80%" | {|style="color:black; border="1" width="80%" | ||
| style="width: 40%;background-color:lightgrey;" | | | style="width: 40%;background-color:lightgrey;" | | ||
| - | ; [[File:Freiburg11_Seq4.png | + | ; [[File:Freiburg11_Seq4.png|400px]] |
| style="width: 40%;background-color:lightgrey;" | | | style="width: 40%;background-color:lightgrey;" | | ||
| - | ; [[File:Freiburg11_Seq5.png | + | ; [[File:Freiburg11_Seq5.png|380px]] |
|} | |} | ||
| Line 111: | Line 117: | ||
{|style="color:black; border="1" width="85%" | {|style="color:black; border="1" width="85%" | ||
| style="width: 100%;background-color:lightgrey;" | | | style="width: 100%;background-color:lightgrey;" | | ||
| - | ;[[File:Freiburg11_Seq6.png | + | ;[[File:Freiburg11_Seq6.png|750px]] |
|} | |} | ||
{{:Team:Freiburg/Templates/footer}} | {{:Team:Freiburg/Templates/footer}} | ||
Revision as of 16:43, 20 September 2011
 "
"
 Contact
Contact