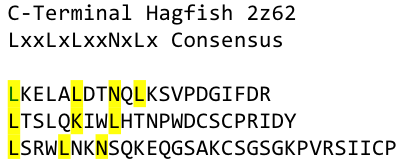Modelling: Rational protein design
The Idea
First there was the idea to replace the chemically produced Ni-NTA columns by some biological, reproducible material. It seemed unreasonable to produce an ion precipitating substance in a complicated, energy consuming way – completely artificial, while so many proteins in nature are capable of the same task – and much more complicated things. But how to harness that?
The second milestone in the generation of the idea came up in a lecture of Prof. Martin, about Nickel allergy. He found a mutation in the TLR-4 receptor that indroduces Histidines in the LRR motif of the protein, which are capable of binding nickel and thus forming complexes of receptors on the cell surface of immune cells, triggering the inflammatory reaction.
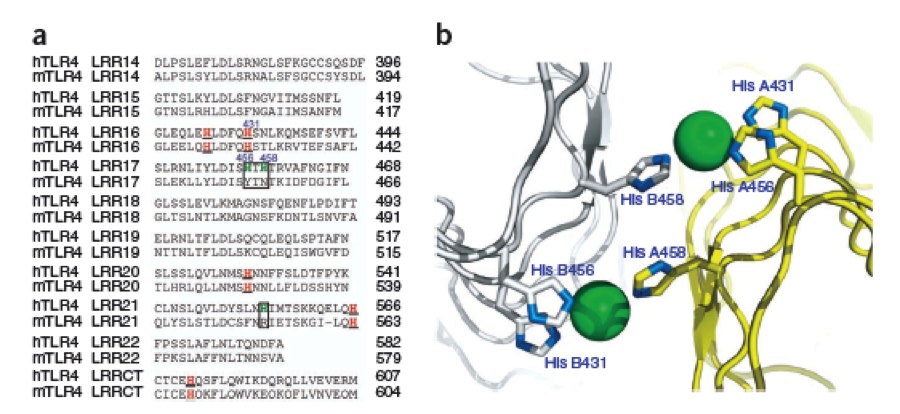
This was the necessary clue that gave answer to the question.
In the following work, a lot of investigation about the perfect Nickel precipitating protein was done. It seemed that there is no optimal natural protein which would serves our needs.
Furthermore, many of the natural LRR proteins – of course – have their own biological function, we we were afraid could interfere in the cellular system we wanted to express them in. Luckily there are over a thousand already known structures of LRR proteins, what gave us a broad choice of motifs to choose and compare.
The Construction of a new protein
After a long and detailed search, we found out that it is most reasonable to use bacterial LRR motifs, since they seemed very well conserved in sequence, and they are the shortest – with only 21 aminoacids per LRR repeat. (Wei 2008, Kajava 1998). We wanted to have the protein as simple and as predictable in behavior and structure as possible.
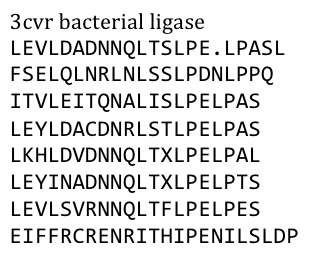
Several search inquiries led to the most conserved and shortest of all bacterial LRR (PDB: NO), unluckily this protein is a toxin derived from Yersinia pestis. We of course did not want to mess around with toxins,
especially not with one of the black plague. We also did not know what amino acid pattern on the LRR motif would cause the toxic effect. There probably would have been no harm from this protein alone, even if we did clone and express it, but why should we do that when we have a huge choice for free at hand anyway. Finally we settled on the bacterial ligase PDB:3CVR. A ligase would be definitively harmless, especially since we planned to restructure it completely.
To do this we needed to understand how the structure of the Ligase was made.
It was that our desired protein LRR motif was only a part of a bigger protein “factory” which included several domains that seemed to serve distinct functions – which we did not want.
So we took only the LRR domain. It looked to us, as if this domain had mostly a structural role in this “factory”, but we could not tell that exactly. To avoid any unspecific biological function it was necessary to rearrange the amino acid composition.
We fed the protein sequence in a free online consensus sequence generator called weblogo from the Berkeley University.
By analyzing the logo it was obvious which positions of the LRR motif were conserved and which not – AND: which of the non conserved aminoacids appeared in what kind of patterns. Were there positions in the protein that required a polar aminoacid? Or non polar, hydrophobic /-philic, charged, non-charged?. We compared the consensus sequence with the 3D structure, using PYMOL, to extract as much information as possible and then came up with this ideal consensus sequence:
The next consideration we had to do, was: how many Nickel do we need on the surface of our ideal Nickel binding protein, in what pattern, with what distances between, and at what angles towards each other to allow proper ion complexation?
Nickel can be complexed by imidazole structures from 4 planar orthogonal directions, as well as to axial positions. It can however only take four ligands at once, preferably in a planar orientation. Cobalt has a bipyramidal setup for ligand-binding, too, but can take up to six ligands. The distances from a N-atom in the Imidazole ring to the ion had to be between 3 and 6 Angström. We found a crystal structure of a different protein (PDB:) which was resolved with three Histidines complexing a Nickel ion for a comparison, as well as some old publications that analyzed peptide ion bonds that taught us how the complex should look like. (Jordan 1974)
We wanted our protein to be short. First, to cause as little unspecific binding as possible, second, to allow an easy expression and third to keep costs los for gene synthesis. For a proper purification of His-tagged proteins in Ni-NTA columns we knew that there are up to three Nickel involved in the interaction between the His tag (which consists of 6 or 7 Histidines) and the column. These have to be in close proximity to one another. There are always two Histidines of the tag binding to one ion.
This was what we had to implement into our LRR backbone.
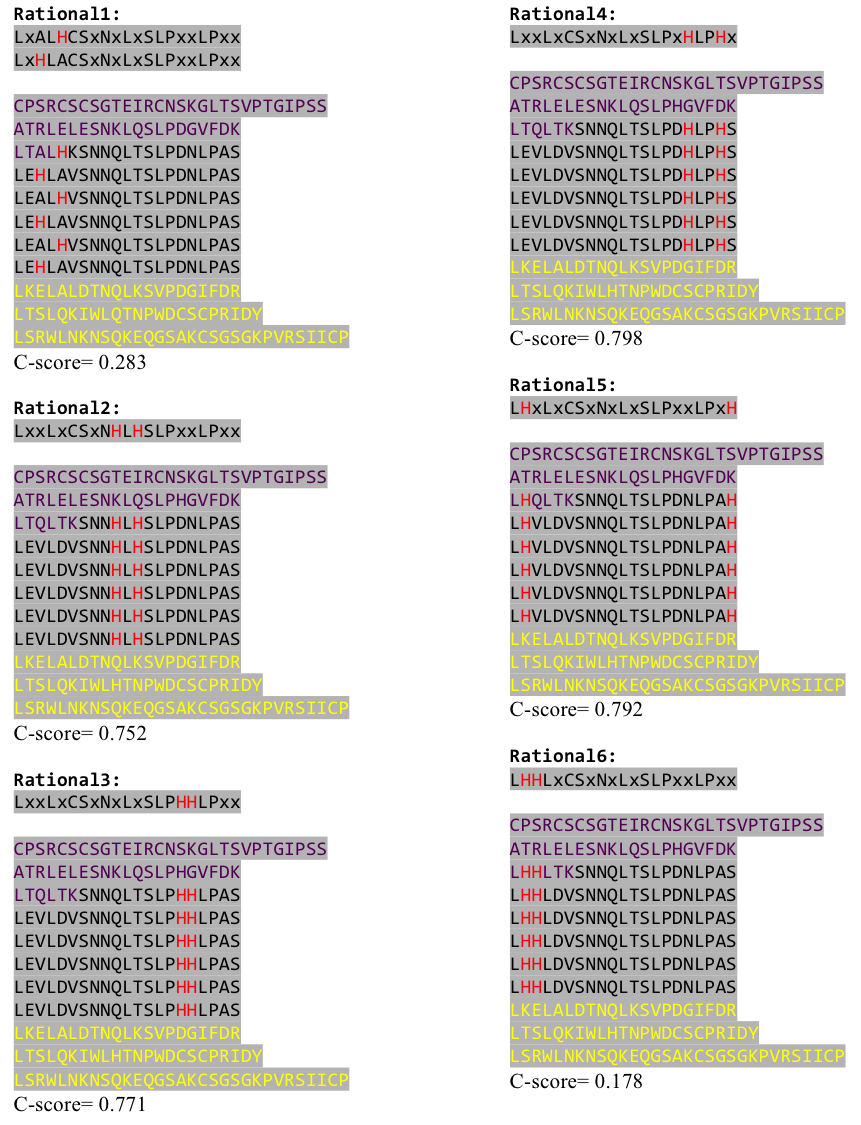
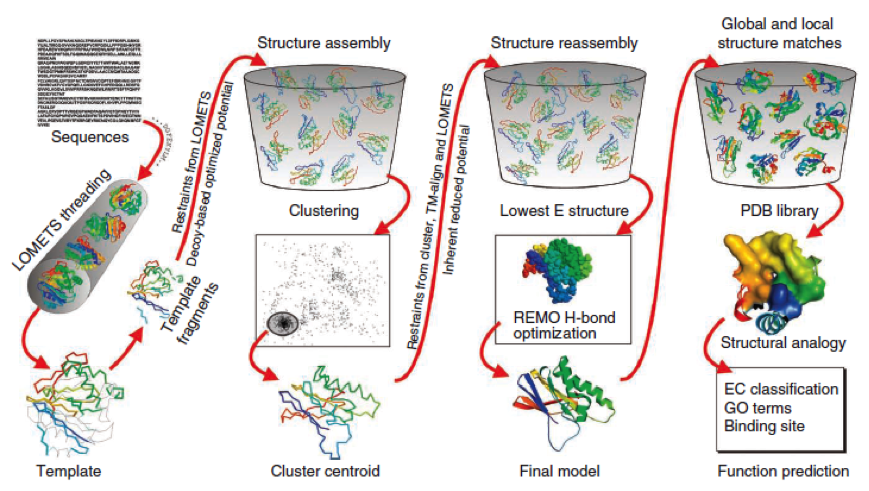
To get a first impression how Histidines are positioned in a bacterial Ligase, we replaced all non conserved aminoacids by Histidines and sent the sequence for a 3D sequence prediction to I-TASSER, an online structure prediction software tool.
| I-Tasser: Structure prediction
|
- I-Tasser is a 3-dimensional protein prediction method which can be used via a downloadable program or a online server. It is provided by the Yang-Zhang lab of the University of Kansas.
The community-wide Critical Assessment of Structure Prediction (CASP) ranked the I-Tasser as the best protein prediction method in the server section.
The method of the I-Tasser can be divided into 4 stages:
Stage 1 is called threading. In this stage the sequence or parts of the sequence or motifs are compared with already known protein structures to identify evolutionary relatives and predict secondary structures. This gives a sequence profile and together with the query sequence it is threaded by LOMETS, a server combining seven threading programs which evaluate the sequence respectively the templates with an individual score. The top templates are selected for further consideration.
Stage 2 is responsible for structural assembly. The structure is modeled ab initio by aligning different templates from the threading. This is a very complex process and different programs are involved.
Stage 3 is a model selection of the achievements of stage 2 with a following refinement of the predicted models.
Stage 4 announces the function of the predicted molecule by its structural conformation comparing with already known proteins of PDB database.
The predicted models are all evaluated by the C-Score, which assesses the quality of the prediction. It has a value from -5 to 2; the more positive the C-Score the merrier and more plausible the predicted structure is.
1.Ambrish Roy, Alper Kucukural, Yang Zhang. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, vol 5, 725-738 (2010).
2.Yang Zhang. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins, vol 69 (Suppl 8), 108-117 (2007).
|
We received this pdb file with a C-score= 0.53. Obviously this was not a realistic model, since the folding would surely be impaired by so many Histidines everywhere. But is was useful to find several good locations, were the Nickel could fit in in respect to the requirements it has for binding ligands.
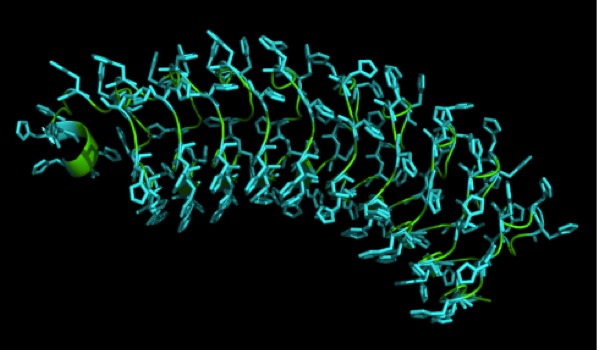
We manually fit in a Nickel ion, as it was crystallized in the mentioned PDB file, and measured the distances and evaluated the 3dimensional orientation of the Histidines towards the Nickel. Histidines can coordinate ligands with their free electron pair pointing planar away from the imidazole ring.
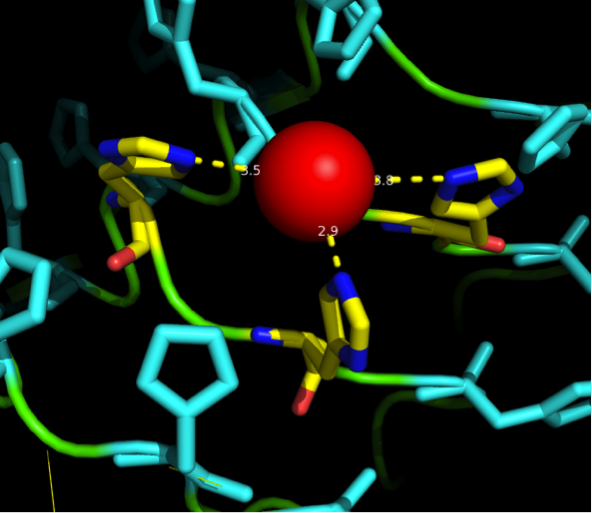
What we further realized from the structure file was, that the end of the LRR segments were “open”, that means, the hydrophobic core of the protein was exposed and as it is visible in the prediction, curled in on one end into a sort of helix. This means the protein folding is not reliable and the structure needs some caps on both ends to stabilize the LRR core motif. A solution to this was shown by Schmidt et al 2010, who crystallized the TLR-4 receptor.
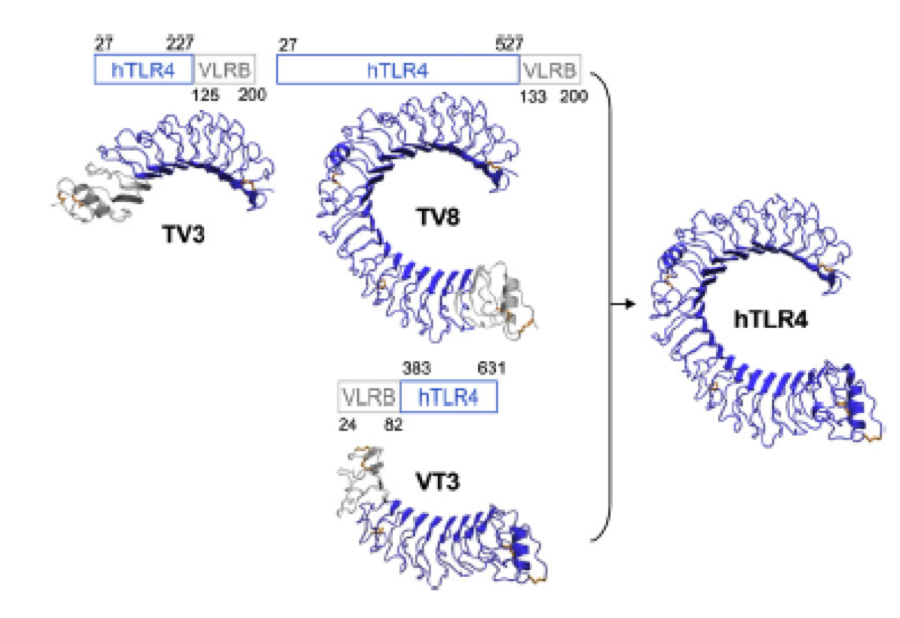 In this very nice piece of work he dissected the TLR4(PDB: 3FXI) into 3 parts, since it was to large and unhandy to be crystallized at once. To overcome this problem of an exposed hydrophobic core, he used the N- and C-terminal protein fragments of a LRR protein derived from hagfish. They tried a variety of different versions of how to glue together his fragments and these N- and C-terminal caps until he found a working one.
We used this knowledge for our purpose and took the same sequences and attached them to our protein sequence. These caps partially still show the typical LRR consensus sequences (which luckily is highly conserved in all kingdoms of life!), which made it possible to fit them onto our stack of LRR loops in the right position.
In this very nice piece of work he dissected the TLR4(PDB: 3FXI) into 3 parts, since it was to large and unhandy to be crystallized at once. To overcome this problem of an exposed hydrophobic core, he used the N- and C-terminal protein fragments of a LRR protein derived from hagfish. They tried a variety of different versions of how to glue together his fragments and these N- and C-terminal caps until he found a working one.
We used this knowledge for our purpose and took the same sequences and attached them to our protein sequence. These caps partially still show the typical LRR consensus sequences (which luckily is highly conserved in all kingdoms of life!), which made it possible to fit them onto our stack of LRR loops in the right position.

To the outer end the protein has a helix on the N-terminal side shielding off the core and the C-terminus the LRR disappears slowly turn by turn with more and more hydrophilic aminoacids replacing the LRR consensus sequence.
After combining these caps with our structural analysis for possible Nickel binding sites and filling the empty positions of the consensus sequence with the ideal sequence derived from the ligase sequence(see above), we were still unsure whether this would work out. To increase our chances of success we designed six different versions with distinct Histidine patterns on the LRR core.
 "
"
 Contact
Contact