Team:Freiburg/Modelling
From 2011.igem.org
(Difference between revisions)
(→Modelling: Rational protein design) |
(→Modelling: Rational protein design) |
||
| Line 5: | Line 5: | ||
First there was the idea to replace the chemically produced Ni-NTA columns by some biological, reproducible material. It seemed unreasonable to produce an ion precipitating substance in a complicated, energy consuming way – completely artificial, while so many proteins in nature are capable of the same task – and much more complicated things. But how to harness that? | First there was the idea to replace the chemically produced Ni-NTA columns by some biological, reproducible material. It seemed unreasonable to produce an ion precipitating substance in a complicated, energy consuming way – completely artificial, while so many proteins in nature are capable of the same task – and much more complicated things. But how to harness that? | ||
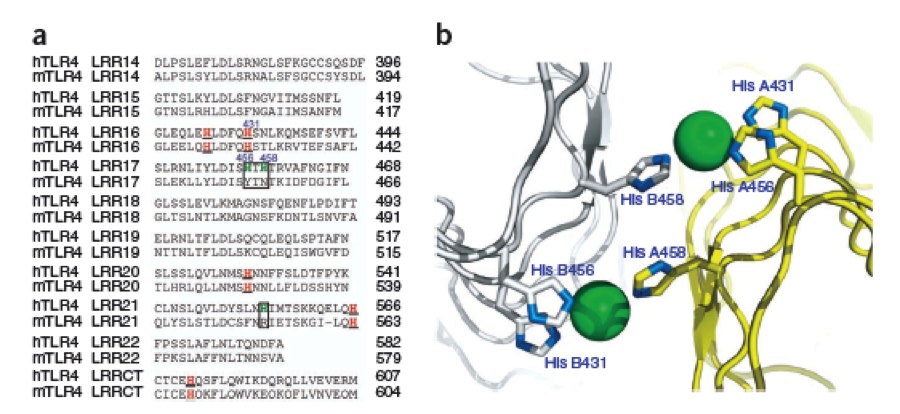
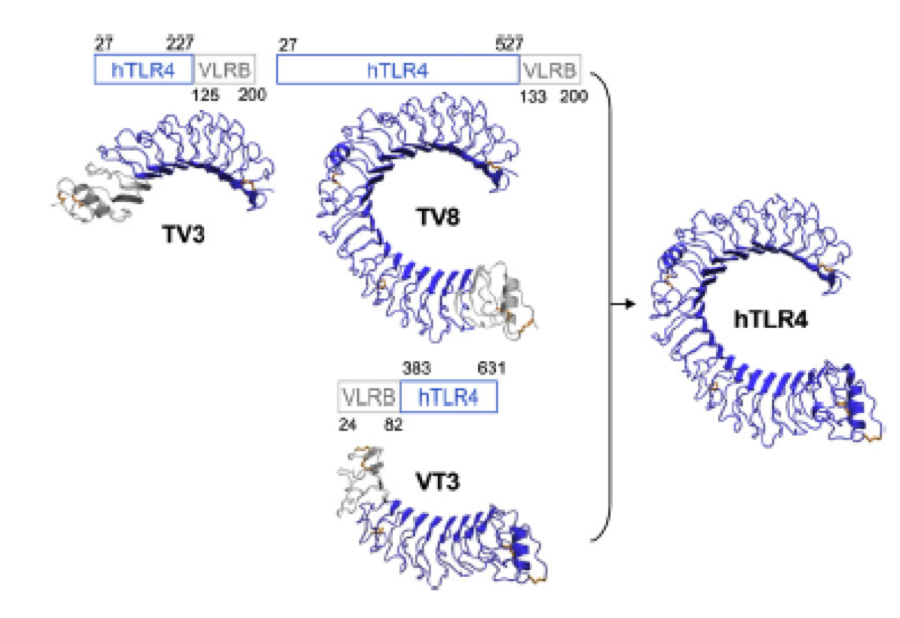
The second milestone in the generation of the idea came up in a lecture of Prof. Martin, about Nickel allergy. He found a mutation in the TLR-4 receptor that indroduces Histidines in the LRR motif of the protein, which are capable of binding nickel and thus forming complexes of receptors on the cell surface of immune cells, triggering the inflammatory reaction. | The second milestone in the generation of the idea came up in a lecture of Prof. Martin, about Nickel allergy. He found a mutation in the TLR-4 receptor that indroduces Histidines in the LRR motif of the protein, which are capable of binding nickel and thus forming complexes of receptors on the cell surface of immune cells, triggering the inflammatory reaction. | ||
| + | |||
[[File:Freiburg11Modelling1.png|border|750px]] | [[File:Freiburg11Modelling1.png|border|750px]] | ||
| + | |||
This was the necessary clue that gave answer to the question. | This was the necessary clue that gave answer to the question. | ||
In the following work, a lot of investigation about the perfect Nickel precipitating protein was done. It seemed that there is no optimal natural protein which would serves our needs. | In the following work, a lot of investigation about the perfect Nickel precipitating protein was done. It seemed that there is no optimal natural protein which would serves our needs. | ||
| Line 17: | Line 19: | ||
So we took only the LRR domain. It looked to us, as if this domain had mostly a structural role in this “factory”, but we could not tell that exactly. To avoid any unspecific biological function it was necessary to rearrange the amino acid composition. | So we took only the LRR domain. It looked to us, as if this domain had mostly a structural role in this “factory”, but we could not tell that exactly. To avoid any unspecific biological function it was necessary to rearrange the amino acid composition. | ||
We fed the protein sequence in a free online consensus sequence generator called weblogo from the Berkeley University. | We fed the protein sequence in a free online consensus sequence generator called weblogo from the Berkeley University. | ||
| + | |||
[[File:Freiburg11Modelling2.png|thumb|750px]] | [[File:Freiburg11Modelling2.png|thumb|750px]] | ||
| Line 28: | Line 31: | ||
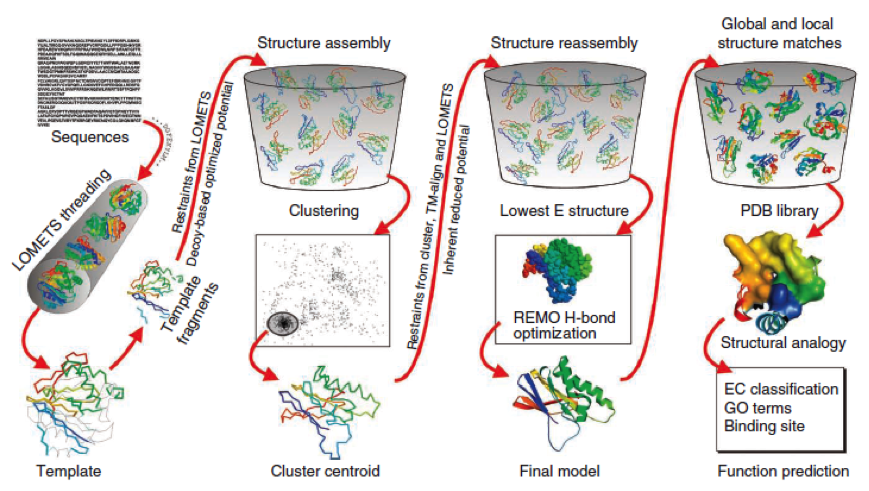
To get a first impression how Histidines are positioned in a bacterial Ligase, we replaced all non conserved aminoacids by Histidines and sent the sequence for a 3D sequence prediction to I-TASSER, an online structure prediction software tool. | To get a first impression how Histidines are positioned in a bacterial Ligase, we replaced all non conserved aminoacids by Histidines and sent the sequence for a 3D sequence prediction to I-TASSER, an online structure prediction software tool. | ||
| + | |||
[[File:Freiburg11_Seq7.png]] | [[File:Freiburg11_Seq7.png]] | ||
| Line 44: | Line 48: | ||
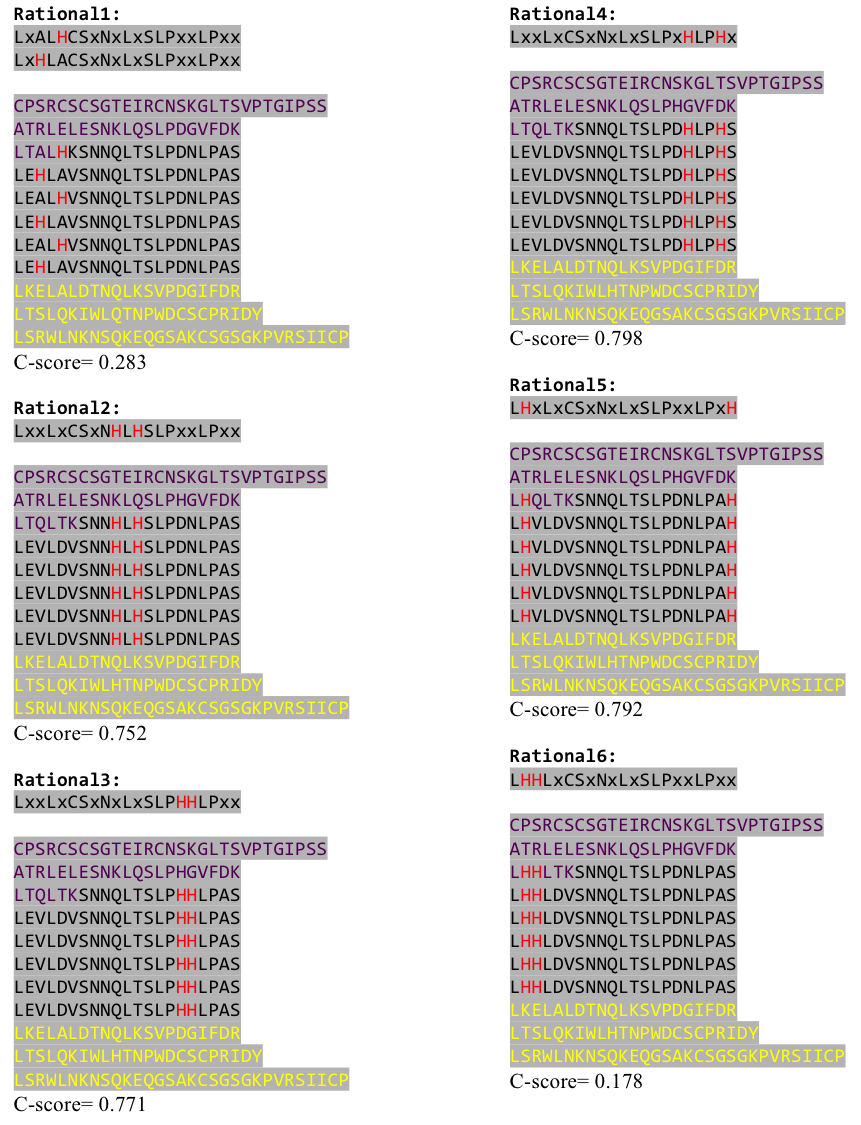
Stage 4 announces the function of the predicted molecule by its structural conformation comparing with already known proteins of PDB database. | Stage 4 announces the function of the predicted molecule by its structural conformation comparing with already known proteins of PDB database. | ||
The predicted models are all evaluated by the C-Score, which assesses the quality of the prediction. It has a value from -5 to 2; the more positive the C-Score the merrier and more plausible the predicted structure is. | The predicted models are all evaluated by the C-Score, which assesses the quality of the prediction. It has a value from -5 to 2; the more positive the C-Score the merrier and more plausible the predicted structure is. | ||
| + | |||
[[File:Freiburg11Modelling3.png|border|750px]] | [[File:Freiburg11Modelling3.png|border|750px]] | ||
| Line 55: | Line 60: | ||
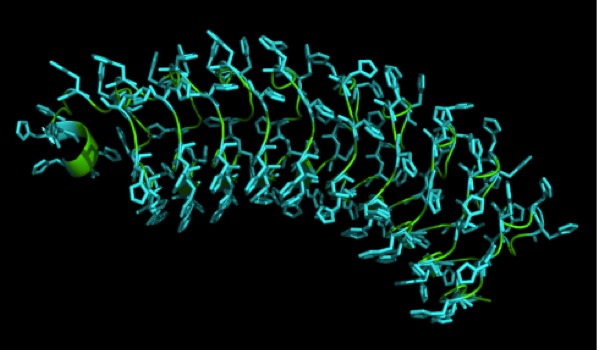
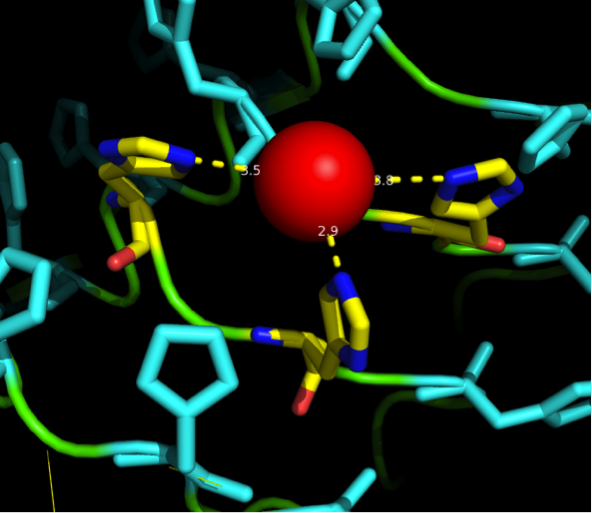
We received this pdb file with a C-score= 0.53. Obviously this was not a realistic model, since the folding would surely be impaired by so many Histidines everywhere. But is was useful to find several good locations, were the Nickel could fit in in respect to the requirements it has for binding ligands. | We received this pdb file with a C-score= 0.53. Obviously this was not a realistic model, since the folding would surely be impaired by so many Histidines everywhere. But is was useful to find several good locations, were the Nickel could fit in in respect to the requirements it has for binding ligands. | ||
| + | |||
[[File:Freiburg11Modelling4.png]] | [[File:Freiburg11Modelling4.png]] | ||
We manually fit in a Nickel ion, as it was crystallized in the mentioned PDB file, and measured the distances and evaluated the 3dimensional orientation of the Histidines towards the Nickel. Histidines can coordinate ligands with their free electron pair pointing planar away from the imidazole ring. | We manually fit in a Nickel ion, as it was crystallized in the mentioned PDB file, and measured the distances and evaluated the 3dimensional orientation of the Histidines towards the Nickel. Histidines can coordinate ligands with their free electron pair pointing planar away from the imidazole ring. | ||
| + | |||
[[File:Freiburg11Modelling5.png]] | [[File:Freiburg11Modelling5.png]] | ||
Revision as of 13:04, 20 September 2011
 "
"
 Contact
Contact