Team:DTU-Denmark/What is sRNA in nature
From 2011.igem.org
(Created page with "{{:Team:DTU-Denmark/Templates/Standard_page_begin|What is: sRNA in nature}} ==sRNA in nature== [[File:DTU1_sRNA_regulatory_circuits.png|330px|thumb|left|Overview of different ''...") |
|||
| Line 1: | Line 1: | ||
| - | {{:Team:DTU-Denmark/Templates/Standard_page_begin| | + | {{:Team:DTU-Denmark/Templates/Standard_page_begin|Background}} |
| - | ==sRNA in nature== | + | ==sRNA in nature == |
[[File:DTU1_sRNA_regulatory_circuits.png|330px|thumb|left|Overview of different '''regulatory circuits which incorporate sRNAs''' in ''E.coli'' and ''Salmonella''. Arrows is activation, bars is repression, and dashed lines indicate indirect regulation. sRNA regulators are shown in red, protein transcription regulators are shown in blue, and target genes and operons are shown in gray. Figure 1 from<span class="superscript">[[#References|[1]]]</span> ]] | [[File:DTU1_sRNA_regulatory_circuits.png|330px|thumb|left|Overview of different '''regulatory circuits which incorporate sRNAs''' in ''E.coli'' and ''Salmonella''. Arrows is activation, bars is repression, and dashed lines indicate indirect regulation. sRNA regulators are shown in red, protein transcription regulators are shown in blue, and target genes and operons are shown in gray. Figure 1 from<span class="superscript">[[#References|[1]]]</span> ]] | ||
Trans-encoded sRNAs are widespread in nature and an increasing number of sRNAs have been found to regulate critical pathways. In prokaryotes, sRNAs are predominantly involved in response to environmental stimuli or stress situations, eg. nutrient starvation, quorum sensing, membrane stress, oxidative stress, and SOS response to DNA damage<span class="superscript">[[#References|[1]]]</span><span class="superscript">[[#References|[2]]]</span>. Examples of well-known sRNAs include: the ''E.coli'' RyhB which downregulates non-essential iron-sulfur containing enzymes when iron is limited and the ''Vibrio'' Qrr which represses quorum sensing when the cell density is low<span class="superscript">[[#References|[3]]]</span>. | Trans-encoded sRNAs are widespread in nature and an increasing number of sRNAs have been found to regulate critical pathways. In prokaryotes, sRNAs are predominantly involved in response to environmental stimuli or stress situations, eg. nutrient starvation, quorum sensing, membrane stress, oxidative stress, and SOS response to DNA damage<span class="superscript">[[#References|[1]]]</span><span class="superscript">[[#References|[2]]]</span>. Examples of well-known sRNAs include: the ''E.coli'' RyhB which downregulates non-essential iron-sulfur containing enzymes when iron is limited and the ''Vibrio'' Qrr which represses quorum sensing when the cell density is low<span class="superscript">[[#References|[3]]]</span>. | ||
Revision as of 11:55, 21 September 2011
Background
sRNA in nature
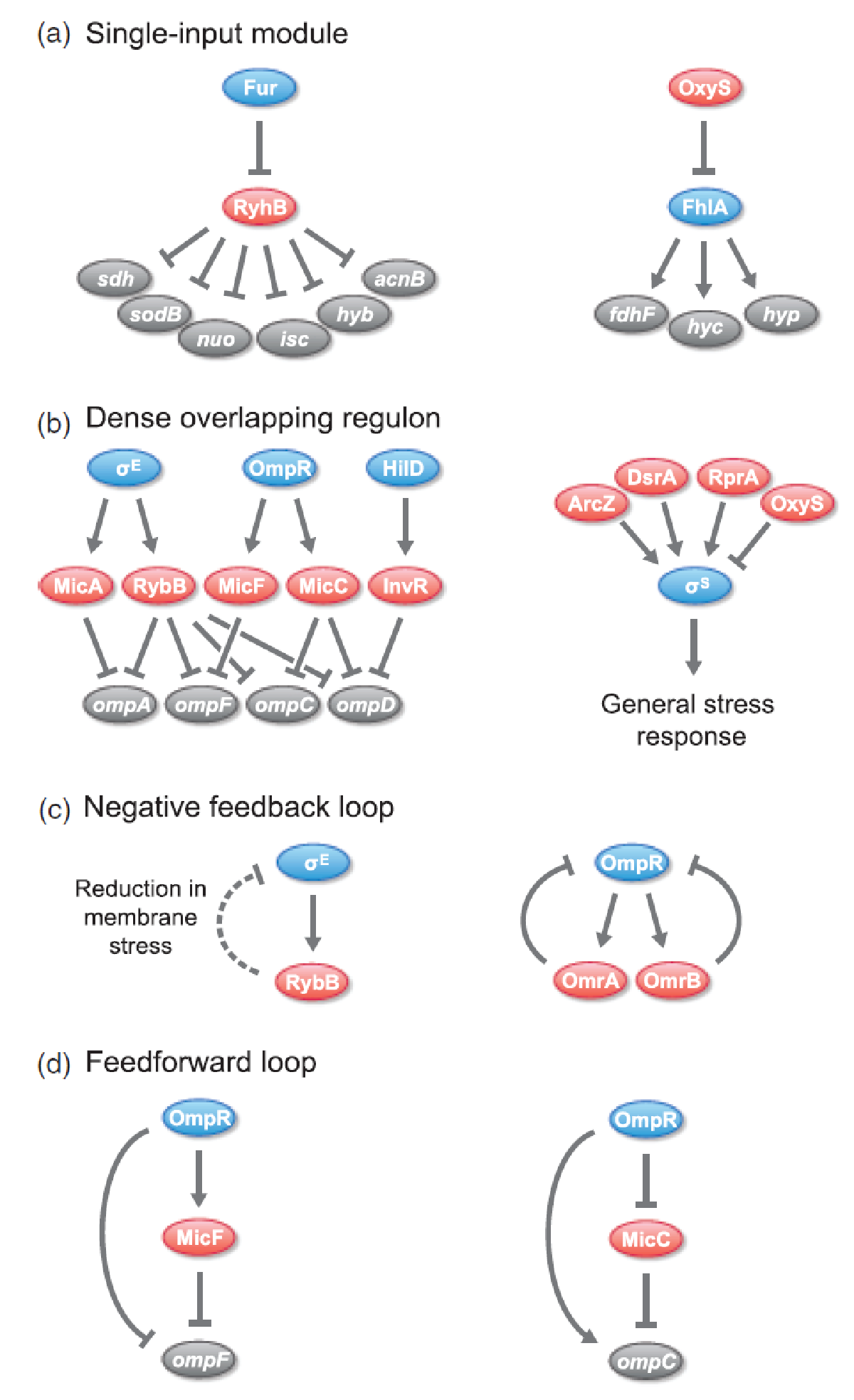
Trans-encoded sRNAs are widespread in nature and an increasing number of sRNAs have been found to regulate critical pathways. In prokaryotes, sRNAs are predominantly involved in response to environmental stimuli or stress situations, eg. nutrient starvation, quorum sensing, membrane stress, oxidative stress, and SOS response to DNA damage[1][2]. Examples of well-known sRNAs include: the E.coli RyhB which downregulates non-essential iron-sulfur containing enzymes when iron is limited and the Vibrio Qrr which represses quorum sensing when the cell density is low[3].
sRNAs have been found to be incorporated in various regulatory circuits which is similar to network motifs found in transcriptional regulatory networks, eg. single-input module (SIM), dense overlapping regulon (DOR), positive feedback loop, negative feedback loop, and feedforward loop (for those geeky guys into gene regulation)[1].
References
[1] Beisel, Chase L., and Gisela Storz. “Base pairing small RNAs and their roles in global regulatory networks.” FEMS Microbiology Reviews 34, no. 5 (2010): 866-882.
[2] Levine, Erel, Zhongge Zhang, Thomas Kuhlman, and Terence Hwa. “Quantitative characteristics of gene regulation by small RNA.” PLoS biology. 5, no. 9 (2007).
[3] Waters, Lauren S., and Gisela Storz. “Regulatory RNAs in Bacteria.” Cell 136, no. 4 (2009): 615-628.
 "
"








