Team:DTU-Denmark/What is Cis versus trans
From 2011.igem.org
(→Cis-acting versus trans-acting sRNA) |
(→) |
||
| Line 6: | Line 6: | ||
| - | + | __NOTOC__ | |
<html></div><div class="whitebox article"></html> | <html></div><div class="whitebox article"></html> | ||
Revision as of 11:50, 21 September 2011
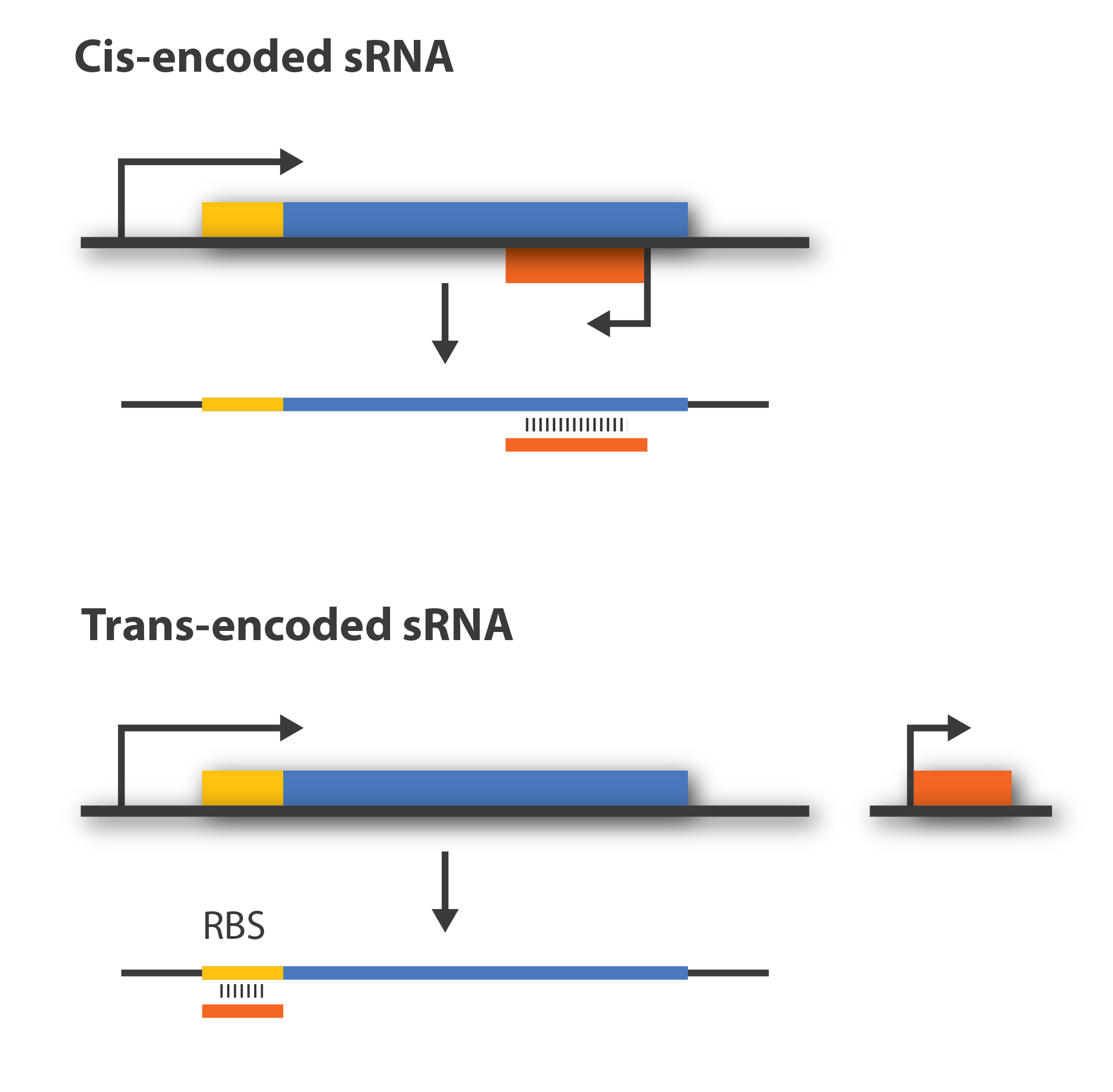
Background: Cis-acting versus trans-acting sRNA
Regulatory sRNAs are divided into different sub-groups depending on their genomic locations. Cis-encoded or antisense sRNAs are encoded just opposite the one gene they regulate whereas trans-encoded sRNAs are located somewhere else in the genome and can aim at multiple number of target mRNA. Antisense RNA share extensive sequence complementarity with its target mRNA and hence do not require the RNA chaperone, Hfq, to stabilize the complex. This is in contrast to trans-encoded sRNAs which often only posses a complementary base-pairing region of 6-20 nucleotides and often require Hfq[1]. We are focusing on trans-encoded sRNA in the project, so whenever we write sRNA we implicit mean trans-encoded sRNA.
References
[1] van Vliet, Arnoud Hm, and Brendan W Wren. “New levels of sophistication in the transcriptional landscape of bacteria.” Genome biology. 10, no. 8 (2009).
 "
"