Team:DTU-Denmark/Notebook
From 2011.igem.org
(Difference between revisions)
(→PChiP/PChiP(mutated)-lacZ fusion) |
(→PTet-chiX fusion) |
||
| Line 41: | Line 41: | ||
#'''plating the bacterial'''and grow over night in 37℃ incubator | #'''plating the bacterial'''and grow over night in 37℃ incubator | ||
#'''start of a liquid culture''' of transformed strain. | #'''start of a liquid culture''' of transformed strain. | ||
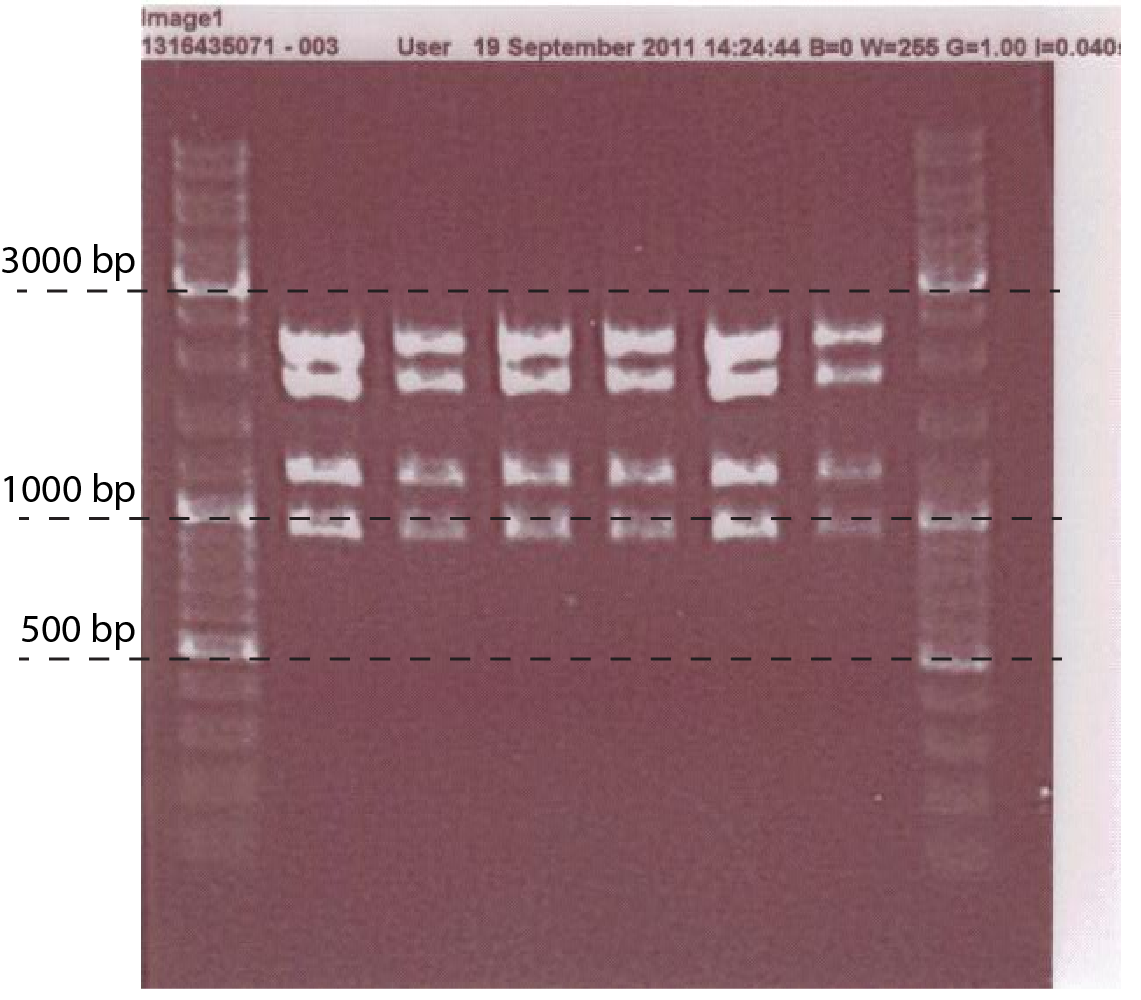
| - | #'''Mini prep the plasmid, do digestion analysis:'''with EcoRI, PstI '''(fig.2)''' | + | #'''Mini prep the plasmid, do digestion analysis:'''with EcoRI, PstI '''(fig.2)''' [[File:DTU-ChiX.png|400px|thumb|left| Fig.2. Colony PCR of cells transformed with P<sub>tet</sub>-chiX inside pSB1C3, CS149 and CS150 primers were used. Expected band size is 1391 bp]] |
#'''subclone Ptet-chiX part into pSB4k5''' | #'''subclone Ptet-chiX part into pSB4k5''' | ||
#'''transformation''' to ''E. coli'' NM522 cells. | #'''transformation''' to ''E. coli'' NM522 cells. | ||
Revision as of 02:15, 22 September 2011
Notebook
Contents |
Constructed plasmids
PChiP/PChiP(mutated)-lacZ fusion
The plasmids containing this DNA fragments were obtained by the following procedure:
- PCR of:
- upstream part of ChiP using E. coli w3110 genomic DNA as a template and TAQ enzyme. Using two different downstream primers (one wild type and one with mutation). we obtained two types of ChiP: original and mutated, which has altered binding site for ChiX.
- lacZ genefrom BBa_I72005 biobrick using phusion enzyme.
- Restriction digestions of:
- ChiP original and ChiP mutated with XbaI and NcoI,
- lacZ gene with BspHI and PstI
- GFP gene with BspHI and PstI
- pSB1C3 with EcoRI and PstI
- ligations of:
- ChiP normal with lacZ and pSB1C3
- ChiP mutated with lacZ and pSB1C3
- transformations to E. coli NM522 cells.
- plating the bacterialand grow over night in 37℃ incubator
- start of a liquid culture of transformed strain.
- Mini prep the plasmid, do digestion analysis:with EcoRI, PstI, EcoRV. (fig.1)
- Transformation to NM522, IG9,IG302.
- Beta-Galactosidase Assay Pick 3 transformants of chip original and chip mutated, and grow. Make betagal assays +/- ara.
PTet-chiX fusion
The plasmids containing this DNA fragments were obtained by the following procedure:
- start liquid culture of minigenes: chiX, chiXR, tetR
- PCR:pSB4k5 (or any other non cam non pSB1 pSB6 plasmid)
- purification of the PCR product
- mini prep minigenes
- Restriction digestions:
- chiX with XbaI and PstI
- tetR with EcoRI and SpaI
- pSB1C3 with EcoRI and PstI
- pSB4k5 with EcoRI and PstI
- ligations of:tetR with chiX and pSB1C3
- transformations to E. coli NM522 cells.
- plating the bacterialand grow over night in 37℃ incubator
- start of a liquid culture of transformed strain.
- Mini prep the plasmid, do digestion analysis:with EcoRI, PstI (fig.2)
- subclone Ptet-chiX part into pSB4k5
- transformation to E. coli NM522 cells.
- Colony PCR with CS149/150 on transformants.Pick 4 positive colonies and grow overnight
- Minipreps the corect ones, Transform into IG302/pSB1C3 PchiPlacZ
- Beta-Galactosidase Assay Pick 2 transformants and grow. Make Beta-Galactosidase Assay +/- atc.
PBad-chiXR fusion
The plasmids containing this DNA fragments were obtained by the following procedure:
- start liquid culture of minigenes: chiXR
- PCR:
- pSB3T5 (or any other non cam non pSB1 pSB6 plasmid)
- pBad
- pSB1C3
- purification of the PCR product
- mini prep minigenes
- Restriction digestions:
- pBad with SpaI and EcoRI
- chiXR with XbaI and PstI
- pSB1C3 with EcoRI and PstI
- pSB4k5 with EcoRI and PstI
- ligations of:pBad with chiXR and pSB1C3
- transformations to E. coli NM522 cells.
- plating the bacterialand grow over night in 37℃ incubator
- start of a liquid culture of transformed strain.
- Mini prep the plasmid, do digestion analysis:with EcoRI, PstI (fig.2)
- subclone PBad-chiXR part into pSB3T5
- transformation to E. coli NM522 cells.
- Colony PCR with CS149/150 on transformants.Pick 4 positive colonies and grow overnight
- Minipreps the corect ones, Transform into NM522/pSB1C3PchiP-lacZ and IG09/pSB1C3PchiP-lacZ
- Beta-Galactosidase Assay Pick 2 transformants and grow. Make betagal assays +/- ara.
Strain construction
The aim of this experimental section was to create strain with following gene deletetions:lacZYA and chbBCARG operon.
- PCR of recombineering linear pieces of DNA using pSLD31 plasmid and RS233/RS234 primers pair (lacZYA deletion) and chb upstream/downstream primers pair. pSLD31, was provided by our supervisor Sebastién Lemire, and it contains a kanamycin resistance gene flanked by lox66 and lox71 sites. The upstream/downstream primers pair anneal at the lox sites and they contain overhangs providing homology region to the sequences flanking "gene-to-be" deleted.
- Transformation of the E. coli W3110 strain containing pSLD18 plasmid (provides Red recombineering system) with pfhc3220 plasmid (containig Cre recombinase).
- Once the strain containing both plasmids was constructed we were ready to proceed with the first round of recombineering. We followed the protocol describing this procedure to achieve first gene deletion - $\Delta$chbBCARG. First we screened for the gain of kanamycin resistance and did the colony PCR to confirm correct insert size. Then the Cre recombinase was activated and facilitated pop-out event of the resistance marker. We screened for loss of resistance and, again, confirmed the correct sizes with colony PCR. Colony PCRs were done using CSchb upstream/downstream primers pair. Correct strain was saved as IG7 and stored under -80oC.
- As the next step, the IG7 strain was transformed with linear piece of DNA designed for lacZYA deletion. Gain of kanamycin resistance was screened for and the expected size of the insert was confirmed with colony PCR. After that the loss of resistance was conducted by growing the cells in minimum medium arabinose (0.15%). We screened for the pop-out event and colony PCRed the strain to confirm the correct recombineering. Primers used were called CSlac upstream/downstream. Finally, the strain was named IG9 and saved under -80oC.
Primers used:
- RS233 ACCAGTACGTTTTCCGCAGCTGTGGGTCAAAGAGGCATGACCTCCTTAGGAATTCTACCG
- RS234 GCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGACTAGCGGATCCTACCGTT
- chb upstream GGCCTGAGTTCTTAATTATCTTCGCGAATTATTTGCCCGATGTGTAGGCTGGAGCTGCTT
- chb downstream CCGGATGCAAGGTTCACGCCGCATCTGGCAAACATCCTCACATATGAATATCCTCCTTAG
- CSchb upstream TATCCGGCCTACAAAACTGTG
- CSchb downstream GCTGAATACGACACTGATCCAC
- CSlac upstream CGCTACAACGGGTAGCAA
- CSlac downstream TCGTTGGGCTGATGATCATAA
PBAD/araC promoter improvement
 "
"