Team:Cambridge/Protocols/PCR
From 2011.igem.org
(→Practice) |
|||
| (32 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Template:Team:Cambridge/ | + | {{Template:Team:Cambridge/CAM_2011_PROTOCOL_HEAD}} |
==Polymerase Chain Reaction (PCR)== | ==Polymerase Chain Reaction (PCR)== | ||
| Line 13: | Line 13: | ||
The reaction works by the annealing of primers to single stranded DNA, which are extended by DNA polymerase. By repeating the process, the section of DNA between the primers is amplified. | The reaction works by the annealing of primers to single stranded DNA, which are extended by DNA polymerase. By repeating the process, the section of DNA between the primers is amplified. | ||
| - | + | During our project, we are using Phusion Hot Start II DNA Polymerase that is characterised by approximately 50-fold lower error rate than ''Thermus aquaticus'' Taq polymerase (the first thermostable polymerase used in PCR). The polymerase, in addition to having 5'→ 3' DNA polymerase activity, also possesses 3'→ 5' exonuclease activity. | |
| + | |||
| + | The Phusion Polymerase allows us to perform Hot Start PCR - a modified version of PCR that avoids non-specific amplification of DNA. The Phusion Polymerase is reversibly bound to specific Affibody protein, which inhibits the DNA polymerase activity at ambient temperatures. Additionally, the Affibody ligand inhibits the 3'→ 5' exonuclease activity of the polymerase, preventing degradation of primers and template DNA during reaction setup. At polymerisation temperatures, the Affibody molecule is released, rendering the polymerase fully active. | ||
===Practice=== | ===Practice=== | ||
| + | |||
| + | ====Reagents used in PCR reaction==== | ||
The PCR reaction with Phusion Hot Start II DNA Polymerase contains the following reagents: | The PCR reaction with Phusion Hot Start II DNA Polymerase contains the following reagents: | ||
| + | <center> | ||
{|border="1px" align="center" style="text-align:center;" | {|border="1px" align="center" style="text-align:center;" | ||
| - | !Name | + | !colwidth="80" | Name |
!50 μl reaction | !50 μl reaction | ||
!20 μl reaction | !20 μl reaction | ||
!Final concentration | !Final concentration | ||
|- | |- | ||
| - | | | + | |Water (reverse osmosis) || add to make 50 μl || add to make 20 μl || |
|- | |- | ||
|5x Phusion Buffer || 10 μl || 4 μl || 1x | |5x Phusion Buffer || 10 μl || 4 μl || 1x | ||
|- | |- | ||
| - | |10mM dNTPs || 1 μl || 0.4 μl || | + | |10mM dNTPs || 1 μl || 0.4 μl || 200 μM each |
|- | |- | ||
| - | |Forward Primer || x μl || x μl || 0.5 μM | + | |Forward Primer 10 μM|| x μl || x μl || 0.5 μM |
|- | |- | ||
| - | |Reverse Primer || x μl || x μl || 0.5 μM | + | |Reverse Primer 10 μM || x μl || x μl || 0.5 μM |
|- | |- | ||
|Template DNA || x μl || x μl || | |Template DNA || x μl || x μl || | ||
| Line 37: | Line 42: | ||
|Phusion Polymerase || 0.5 μl || 0.2 μl || 0.02 U/μl | |Phusion Polymerase || 0.5 μl || 0.2 μl || 0.02 U/μl | ||
|} | |} | ||
| - | + | </center> | |
'''Tip:''' The recommended buffer is 5x Phusion HF Buffer, used as a default buffer for high fidelity amplification. | '''Tip:''' The recommended buffer is 5x Phusion HF Buffer, used as a default buffer for high fidelity amplification. | ||
| - | '''Tip:''' When you receive primers synthesized by a biotechnological company | + | '''Tip:''' The DNA polymerase should be pipetted carefully and gently as the high glycerol content (50%) in the storage buffer may otherwise lead to pipetting errors. |
| - | *centrifuge tubes prior to opening to prevent loss of pelleted oligonucleotides | + | |
| - | *add the amount of water specified in the table provided with primers to obtain 100 μM solution | + | When you receive primers synthesized by a biotechnological company |
| + | *centrifuge tubes prior to opening to prevent loss of pelleted oligonucleotides (at 3000 rpm for 1 minute) | ||
| + | *add the amount of deionized water specified in the table provided with primers to obtain 100 μM solution | ||
*prepare a 4-fold dilution of a portion of the primer solution | *prepare a 4-fold dilution of a portion of the primer solution | ||
Only the primers and the DNA template are specific to the reaction. The remaining reagents can be made as a 'Master Mix' in order to reduce the need to repeatedly pipette small volumes which amplify experimental error. | Only the primers and the DNA template are specific to the reaction. The remaining reagents can be made as a 'Master Mix' in order to reduce the need to repeatedly pipette small volumes which amplify experimental error. | ||
| + | |||
| + | The amount of template DNA added to a PCR reaction varies with its complexity: | ||
| + | *for plasmid, lambda or BAC DNA, 1 pg - 10 ng per 50 μl reaction volume is recommended | ||
| + | *for high complexity genomic DNA, the amount of DNA template should be 50 - 250 ng per 50 μl reaction volume. | ||
As with all enzyme-containing reagents, the master mix should be kept in the freezer and small aliquots thawed when required. | As with all enzyme-containing reagents, the master mix should be kept in the freezer and small aliquots thawed when required. | ||
| Line 51: | Line 62: | ||
The remainder of the reaction is handled automatically by a PCR machine. It is common practice to perform a [[Team:Cambridge/Protocols/Gel_Electrophoresis | gel electrophoresis]] to extract the correct DNA after PCR. | The remainder of the reaction is handled automatically by a PCR machine. It is common practice to perform a [[Team:Cambridge/Protocols/Gel_Electrophoresis | gel electrophoresis]] to extract the correct DNA after PCR. | ||
| + | ====Conditions of PCR reaction==== | ||
An example of conditions we used for a PCR reaction using Phusion polymerase are as follows: | An example of conditions we used for a PCR reaction using Phusion polymerase are as follows: | ||
| + | <center> | ||
{| border="1" align="center" style="text-align:center;" | {| border="1" align="center" style="text-align:center;" | ||
| - | + | !colspan="2" | Step | |
| - | + | !Temp. | |
| - | |Step | + | !Time |
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | |scope="col" width="100" | '''Hold 1''' |
| - | |1 | + | | |
| - | | | + | |95°C |
| - | | | + | |6 min |
| - | | | + | |
|- | |- | ||
| - | |rowspan= | + | |scope="col" width="100" rowspan="3" | '''Cycling ×30''' |
| - | | | + | |scope="col" width="100" | ''Denaturing'' |
| - | | | + | |scope="col" width="80" | 98°C |
| - | | | + | |scope="col" width="80" | 10 s |
| - | |10 | + | |
|- | |- | ||
| - | | | + | |scope="col" width="80" | ''Annealing'' |
| - | | | + | |scope="col" width="50" | 55°C |
| - | |30 | + | |scope="col" width="50" | 30 s |
|- | |- | ||
| - | | | + | |scope="col" width="80" | ''Elongation'' |
| - | | | + | |scope="col" width="50" | 72°C |
| - | | | + | |scope="col" width="50" | 180 s |
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |50 | + | |
| - | + | ||
| - | + | ||
| - | | | + | |
|} | |} | ||
| + | </center> | ||
| + | |||
| + | '''Tip:''' Use 15-30 s/kb for extension. Do not exceed 1 min/kb. | ||
| + | |||
| + | ===PCR Modifications=== | ||
| + | ====Gradient PCR==== | ||
| + | Gradient PCR is a modification to a standard PCR procedure that allows to optimize an annealing temperature in order to increase the specificity of the amplification process. It is usually conducted in a thermal cycler in which different annealing temperatures can be set in different parts of the block, maintaining constant denaturing and elongation temperatures. Thus, after running a gel with PCR products and comparing the pattern of bands, it is possible to empirically determine the optimal annealing temperature for a given set of primers. | ||
===Safety=== | ===Safety=== | ||
| Line 97: | Line 101: | ||
<br style="clear:both"> | <br style="clear:both"> | ||
| - | {{Template:Team:Cambridge/ | + | {{Template:Team:Cambridge/CAM_2011_PROTOCOL_FOOT}} |
Latest revision as of 20:19, 21 September 2011
Contents |
Polymerase Chain Reaction (PCR)
The goal of PCR is to amplify a section of DNA of interest for DNA analysis (e.g. gene insertion, sequencing, etc). The amplification rate is exponential.
Theory
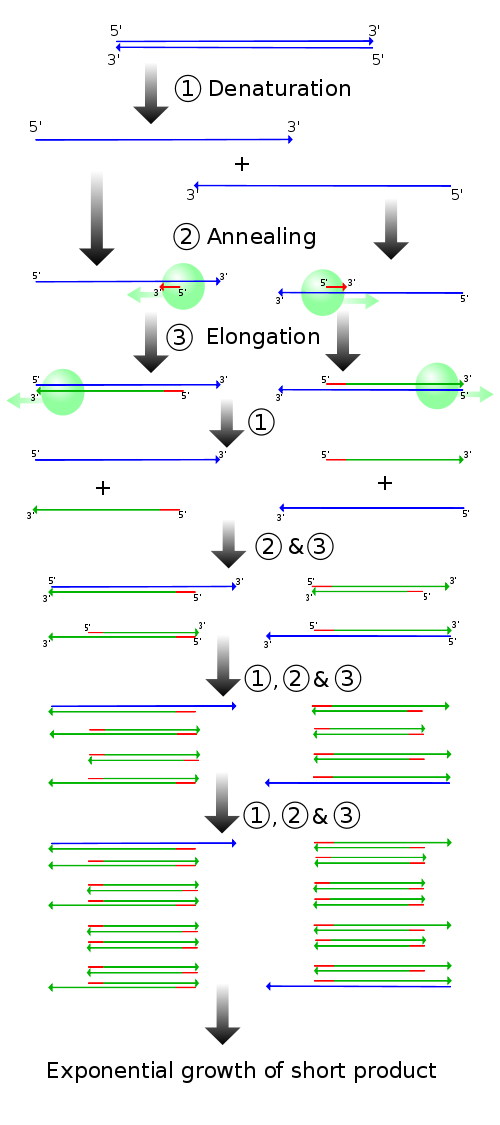
(1) Denaturing at 94–96 °C
(2) Annealing at ~65 °C
(3) Elongation at 72 °C Four cycles are shown here. The blue lines represent the DNA template to which primers ( red arrows ) anneal that are extended by the DNA polymerase ( light green circles ), to give shorter DNA products ( green lines ), which themselves are used as templates as PCR progresses. Credit, WikiMediaThe reaction works by the annealing of primers to single stranded DNA, which are extended by DNA polymerase. By repeating the process, the section of DNA between the primers is amplified.
During our project, we are using Phusion Hot Start II DNA Polymerase that is characterised by approximately 50-fold lower error rate than Thermus aquaticus Taq polymerase (the first thermostable polymerase used in PCR). The polymerase, in addition to having 5'→ 3' DNA polymerase activity, also possesses 3'→ 5' exonuclease activity.
The Phusion Polymerase allows us to perform Hot Start PCR - a modified version of PCR that avoids non-specific amplification of DNA. The Phusion Polymerase is reversibly bound to specific Affibody protein, which inhibits the DNA polymerase activity at ambient temperatures. Additionally, the Affibody ligand inhibits the 3'→ 5' exonuclease activity of the polymerase, preventing degradation of primers and template DNA during reaction setup. At polymerisation temperatures, the Affibody molecule is released, rendering the polymerase fully active.
Practice
Reagents used in PCR reaction
The PCR reaction with Phusion Hot Start II DNA Polymerase contains the following reagents:
| Name | 50 μl reaction | 20 μl reaction | Final concentration |
|---|---|---|---|
| Water (reverse osmosis) | add to make 50 μl | add to make 20 μl | |
| 5x Phusion Buffer | 10 μl | 4 μl | 1x |
| 10mM dNTPs | 1 μl | 0.4 μl | 200 μM each |
| Forward Primer 10 μM | x μl | x μl | 0.5 μM |
| Reverse Primer 10 μM | x μl | x μl | 0.5 μM |
| Template DNA | x μl | x μl | |
| Phusion Polymerase | 0.5 μl | 0.2 μl | 0.02 U/μl |
Tip: The recommended buffer is 5x Phusion HF Buffer, used as a default buffer for high fidelity amplification.
Tip: The DNA polymerase should be pipetted carefully and gently as the high glycerol content (50%) in the storage buffer may otherwise lead to pipetting errors.
When you receive primers synthesized by a biotechnological company
- centrifuge tubes prior to opening to prevent loss of pelleted oligonucleotides (at 3000 rpm for 1 minute)
- add the amount of deionized water specified in the table provided with primers to obtain 100 μM solution
- prepare a 4-fold dilution of a portion of the primer solution
Only the primers and the DNA template are specific to the reaction. The remaining reagents can be made as a 'Master Mix' in order to reduce the need to repeatedly pipette small volumes which amplify experimental error.
The amount of template DNA added to a PCR reaction varies with its complexity:
- for plasmid, lambda or BAC DNA, 1 pg - 10 ng per 50 μl reaction volume is recommended
- for high complexity genomic DNA, the amount of DNA template should be 50 - 250 ng per 50 μl reaction volume.
As with all enzyme-containing reagents, the master mix should be kept in the freezer and small aliquots thawed when required.
The remainder of the reaction is handled automatically by a PCR machine. It is common practice to perform a gel electrophoresis to extract the correct DNA after PCR.
Conditions of PCR reaction
An example of conditions we used for a PCR reaction using Phusion polymerase are as follows:
| Step | Temp. | Time | |
|---|---|---|---|
| Hold 1 | 95°C | 6 min | |
| Cycling ×30 | Denaturing | 98°C | 10 s |
| Annealing | 55°C | 30 s | |
| Elongation | 72°C | 180 s | |
Tip: Use 15-30 s/kb for extension. Do not exceed 1 min/kb.
PCR Modifications
Gradient PCR
Gradient PCR is a modification to a standard PCR procedure that allows to optimize an annealing temperature in order to increase the specificity of the amplification process. It is usually conducted in a thermal cycler in which different annealing temperatures can be set in different parts of the block, maintaining constant denaturing and elongation temperatures. Thus, after running a gel with PCR products and comparing the pattern of bands, it is possible to empirically determine the optimal annealing temperature for a given set of primers.
Safety
No bacteria are used during the reaction there is therefore little or no biological hazard. Nevertheless, it is important to observe correct laboratory procedure and wear appropriate clothing and gloves. PCR occurs at high temperature, and this may present a risk, depending on the PCR machine employed.
Back to Protocols
 "
"
