Team:Cambridge/Project/In Vitro
From 2011.igem.org
(→Thin Films) |
|||
| (38 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Team:Cambridge/CAM_2011_TEMPLATE_HEAD}} | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_HEAD}} | ||
| + | =Isolating Reflectin and Making Thin Films= | ||
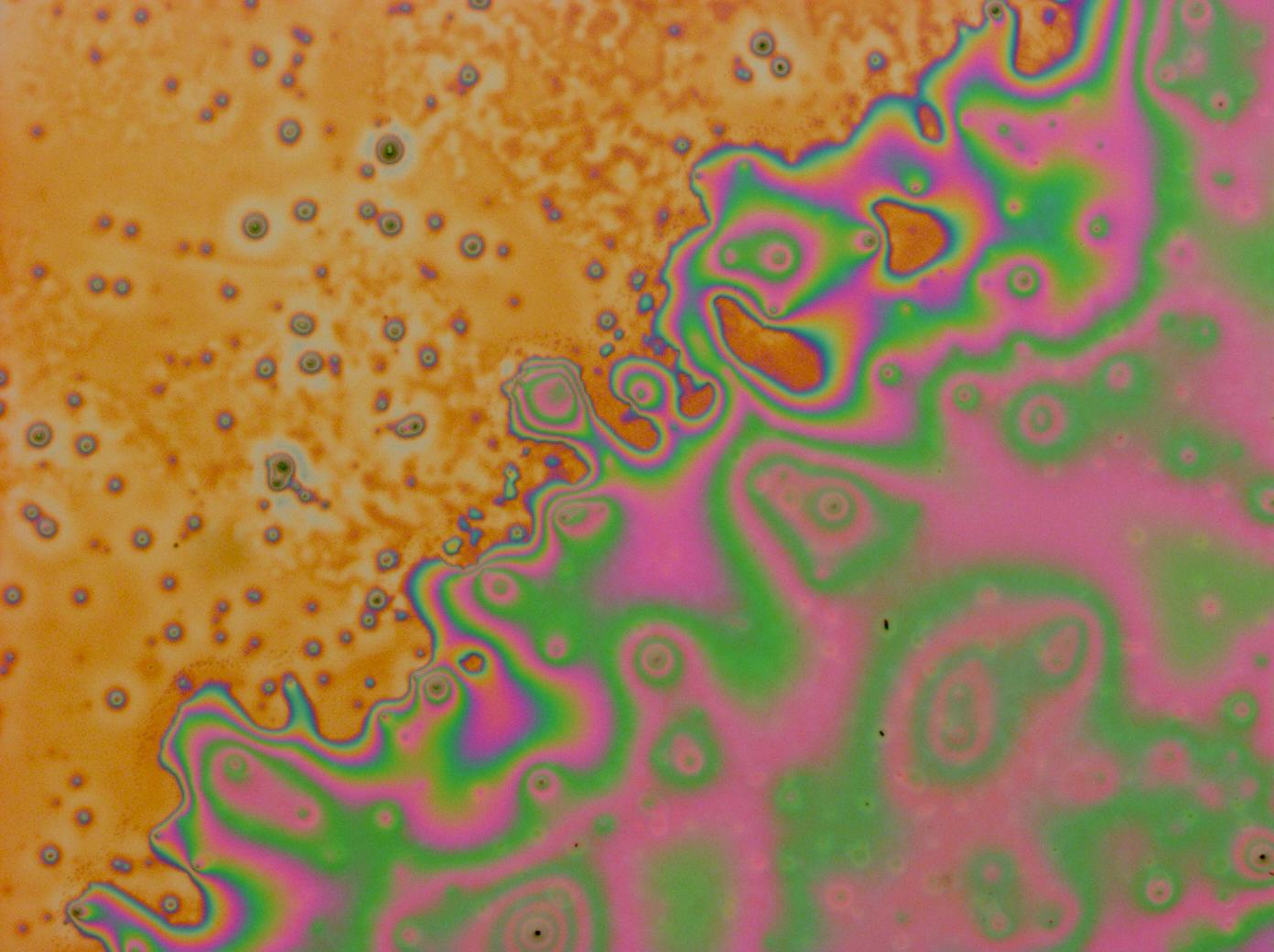
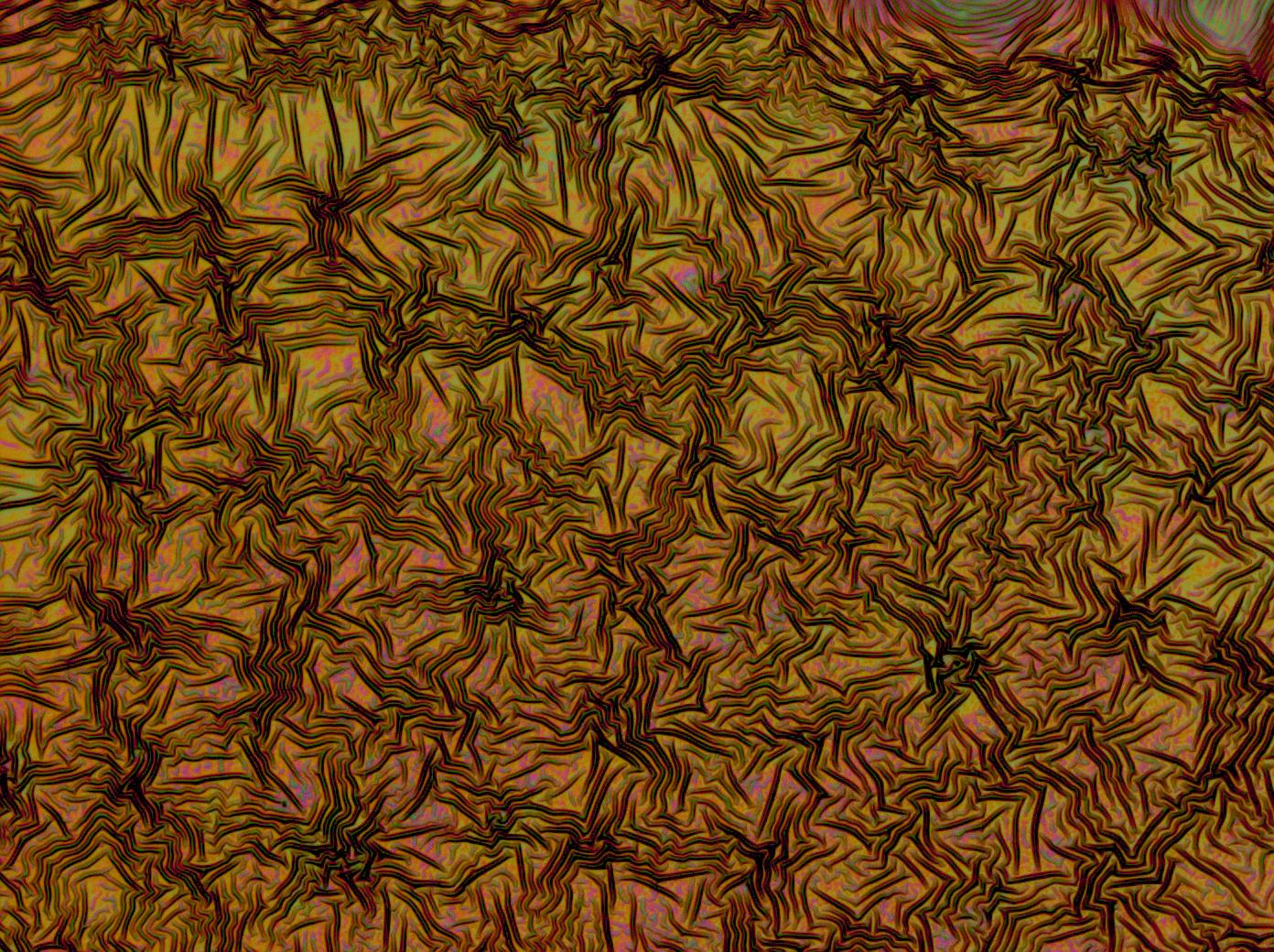
| - | + | [[File:Cam crazy multilayer3.jpg | thumb | 250px | right | A PDMS on reflectin multilayer thin film]] | |
| - | + | Previous in vitro investigations of reflectin showed that it was possible to use a method called [[Team:Cambridge/Protocols/Flow_coating | flow coating]] to deposit a thin layer of reflectin onto a silicon substrate which would then demonstrate structural colour. | |
| - | [[ | + | We wanted to investigate using a different coating method – [[Team:Cambridge/Protocols/Spin_Coating | spin coating]] – and to try out different methods for [[Team:Cambridge/Protocols/Protein_Purification | protein purification]]. A polyhis-tagged variant of the protein was overexpressed and purified using a polyhis affinity column. Thin films were made by precipitating with either [[Team:Cambridge/Protocols/Acetone_Precipitation_of_Proteins | acetone]] or [[Team:Cambridge/Protocols/Ethanol_Precipitation_of_Proteins | ethanol]] and resuspending in HFIP before either spin coating or flow coating onto a silicon wafer. |
| - | + | We also investigated an [[Team:Cambridge/Protocols/Inclusion_Body_Prep | inclusion body prep]], as we found that the protein formed inclusion bodies upon over-expression. By varying our purification and coating methods, we were able to create vibrant thin films. | |
| - | + | ==[[Team:Cambridge/Experiments/Thin_Films | Thin Films]]== | |
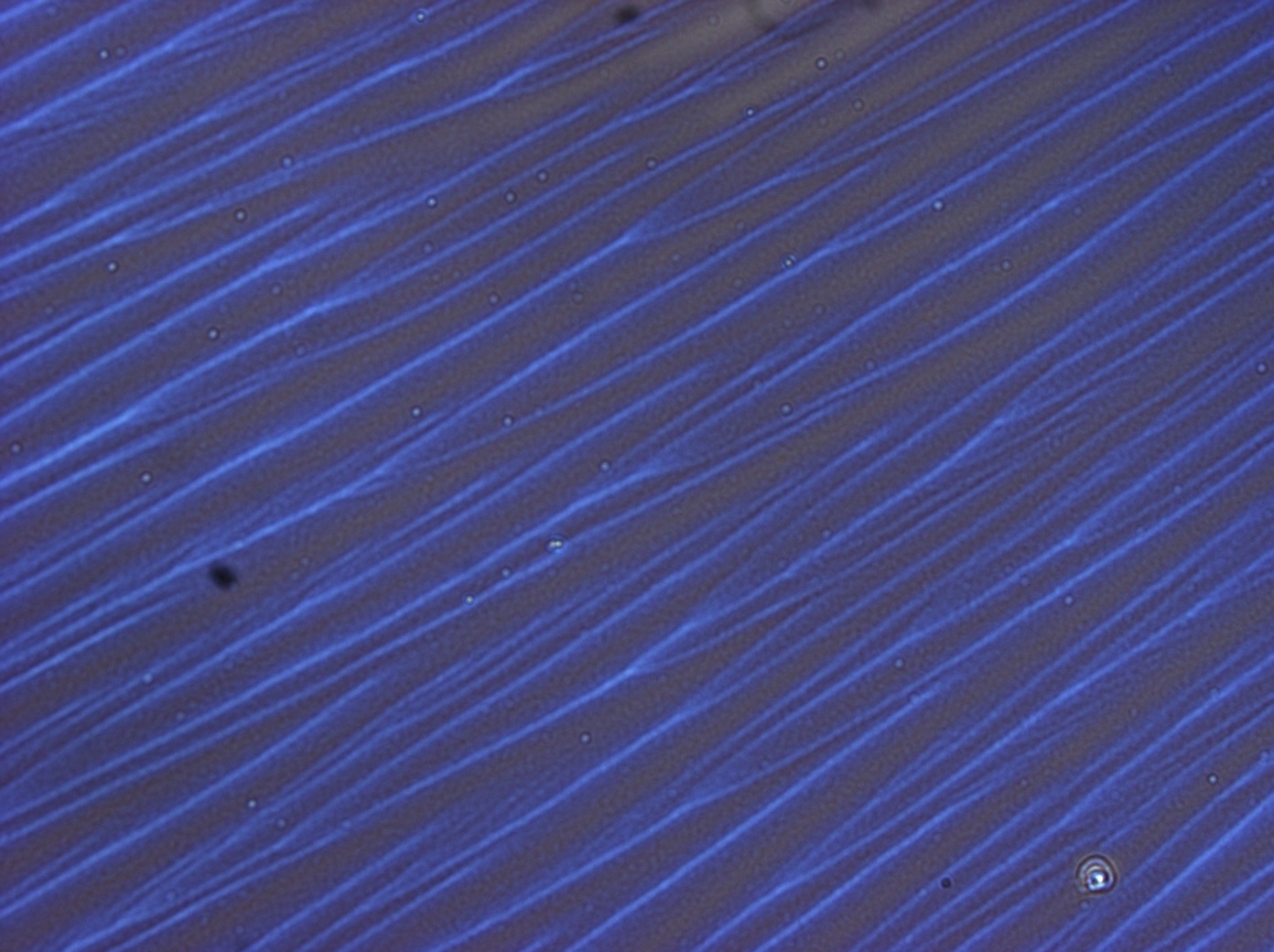
| + | In order to demonstrate structural colour, thin films were created by re-suspending the purified protein in HFIP and [[Team:Cambridge/Protocols/Spin_Coating | spin coating]] or [[Team:Cambridge/Protocols/Flow_coating | flow coating]] the re-suspended protein onto a silicon substrate. This gave us brightly coloured thin films which responded to water vapour in the air by changing colour. We performed controls by making HFIP films and films with Bovine Serum Albumin (BSA, a generic protein) neither of which showed any structural colour. | ||
| - | == | + | <center> |
| + | <gallery widths=150px caption='An initial thin film (left), alongside its controls'> | ||
| + | File:Cam Reflectin Thin Film2.jpg | ''Reflectin HFIP solution showed iridescence after coating'' | ||
| + | File:Cam HFIP only control thinfilm.jpg | ''HFIP (solvent) only control does not exhibit iridescence'' | ||
| + | File:BSAcontrolfilm1.jpg | ''Bovine Serum Albumin makes a dull, striated thin film'' | ||
| + | </gallery> | ||
| + | </center> | ||
| - | + | Our initial films were not uniform in colour and would form crystals when allowed to dry out. We improved this by increasing the purity of our protein as well as refining our coating technique. | |
| - | + | We first tried to reduce the urea content of our sample, in the belief that this would reduce the tendency to crystallise, however, this did not work as our sample became too dilute. Almost by accident, we tried using a [[Team:Cambridge/Experiments/Reflectin_Thin_Films_IV | lower volume of protein]] and found - to our great surprise - that the quality of the thin films produced increased greatly, and the crystallisation was greatly reduced. | |
| - | + | ||
| - | The Norgen Proteospin inclusion body prep | + | The reflectin thin films were dynamic - they responded to water vapour in the air. The video below shows the effect of breathing several times on a thin film - the colour change is quite dramatic. |
| - | + | ||
| + | <center> | ||
| + | <html><iframe width="425" height="349" src="http://www.youtube.com/embed/FoigFqztElE?hl=en&fs=1" frameborder="0" allowfullscreen></iframe></html> | ||
| + | </center> | ||
| + | |||
| + | '''(See more videos of our thin films [http://www.youtube.com/user/cambridgeigem2011 here]).''' | ||
| + | |||
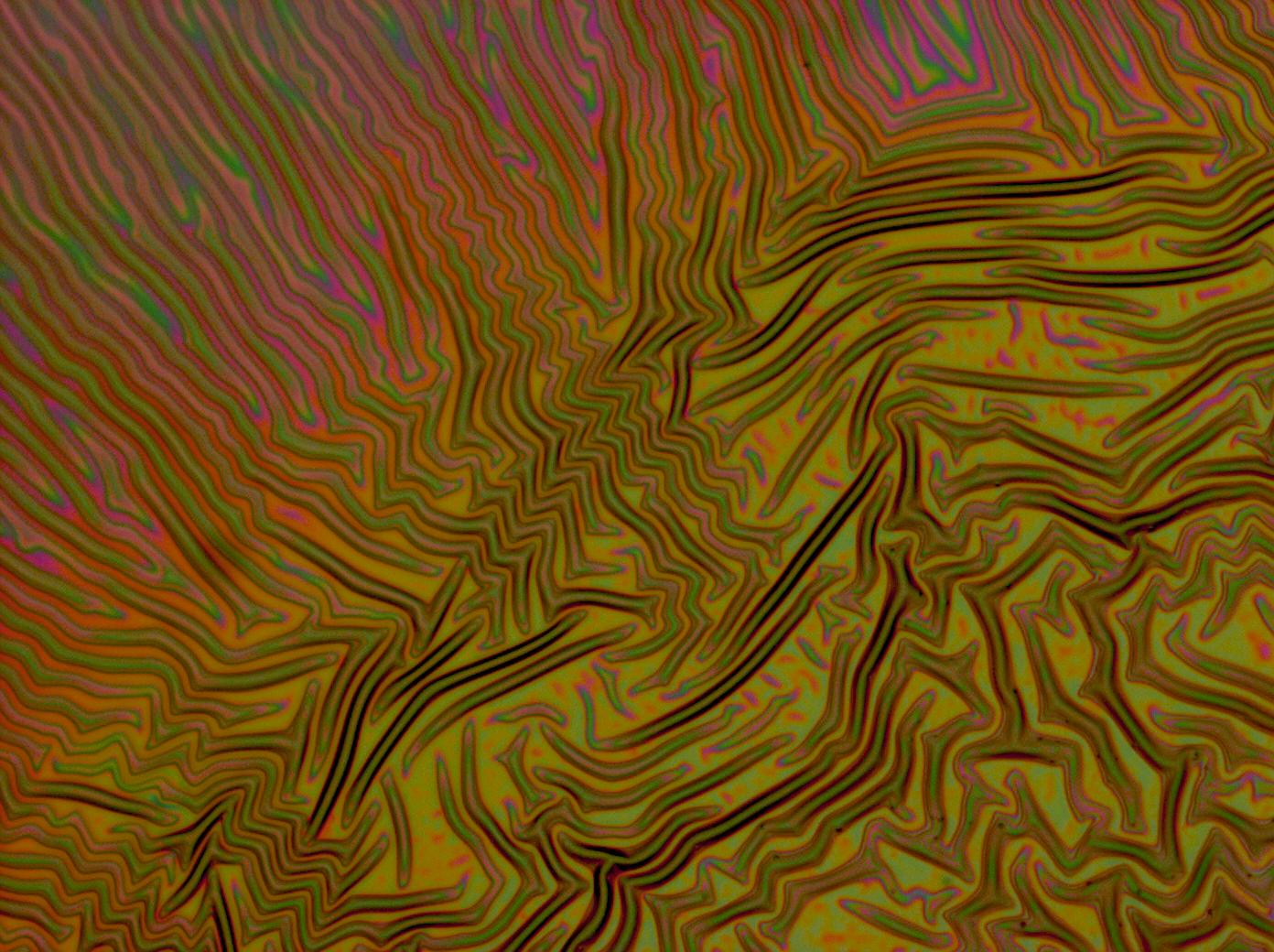
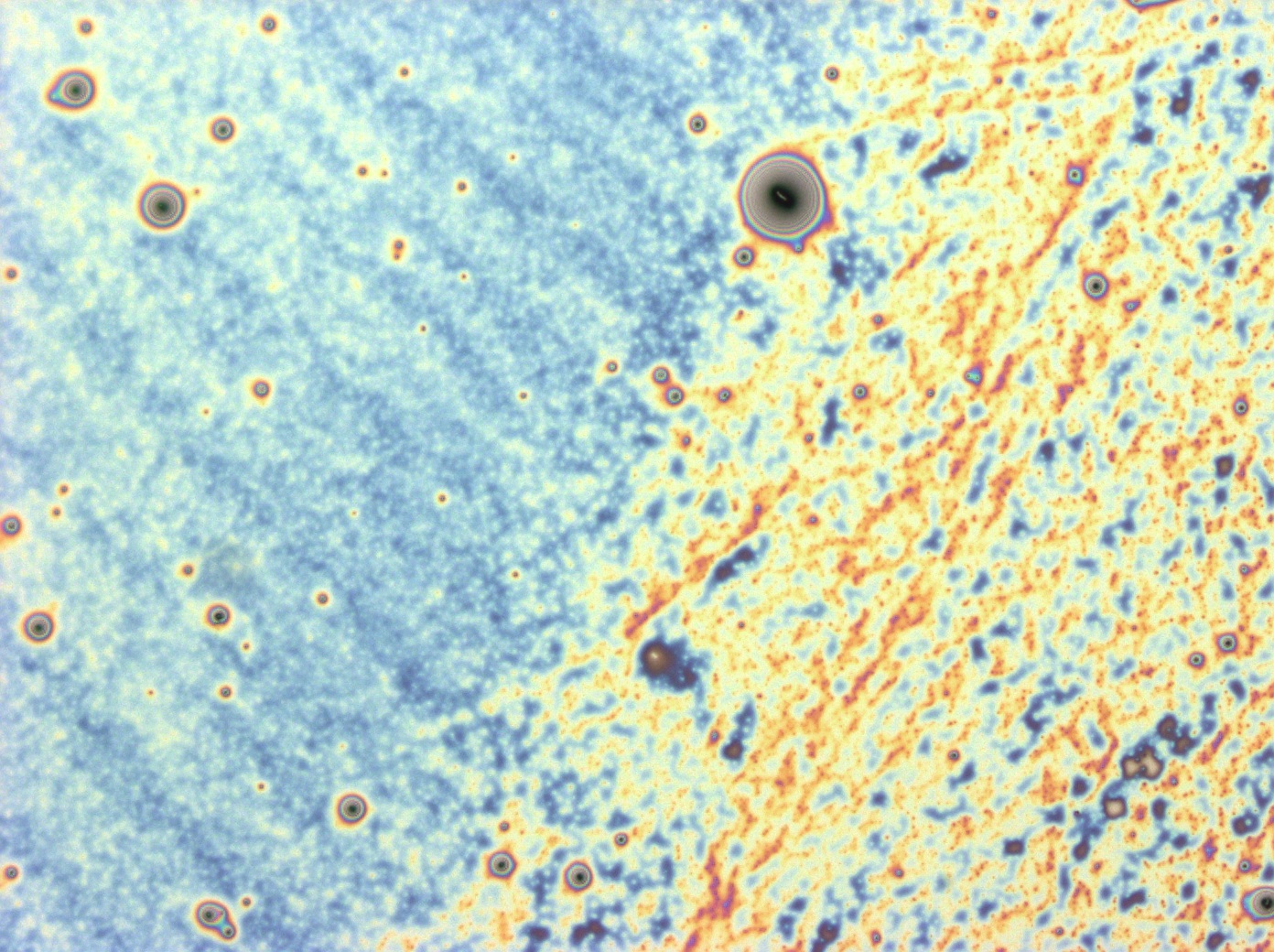
| + | Flushed with success, we then pushed on to make [[Team:Cambridge/Experiments/Reflectin_Thin_Films_VI | multi layers]] of Reflectin, hoping that this would again increase the brightness and uniformity of the thin films. The beauty of the results was astonishing. | ||
| + | |||
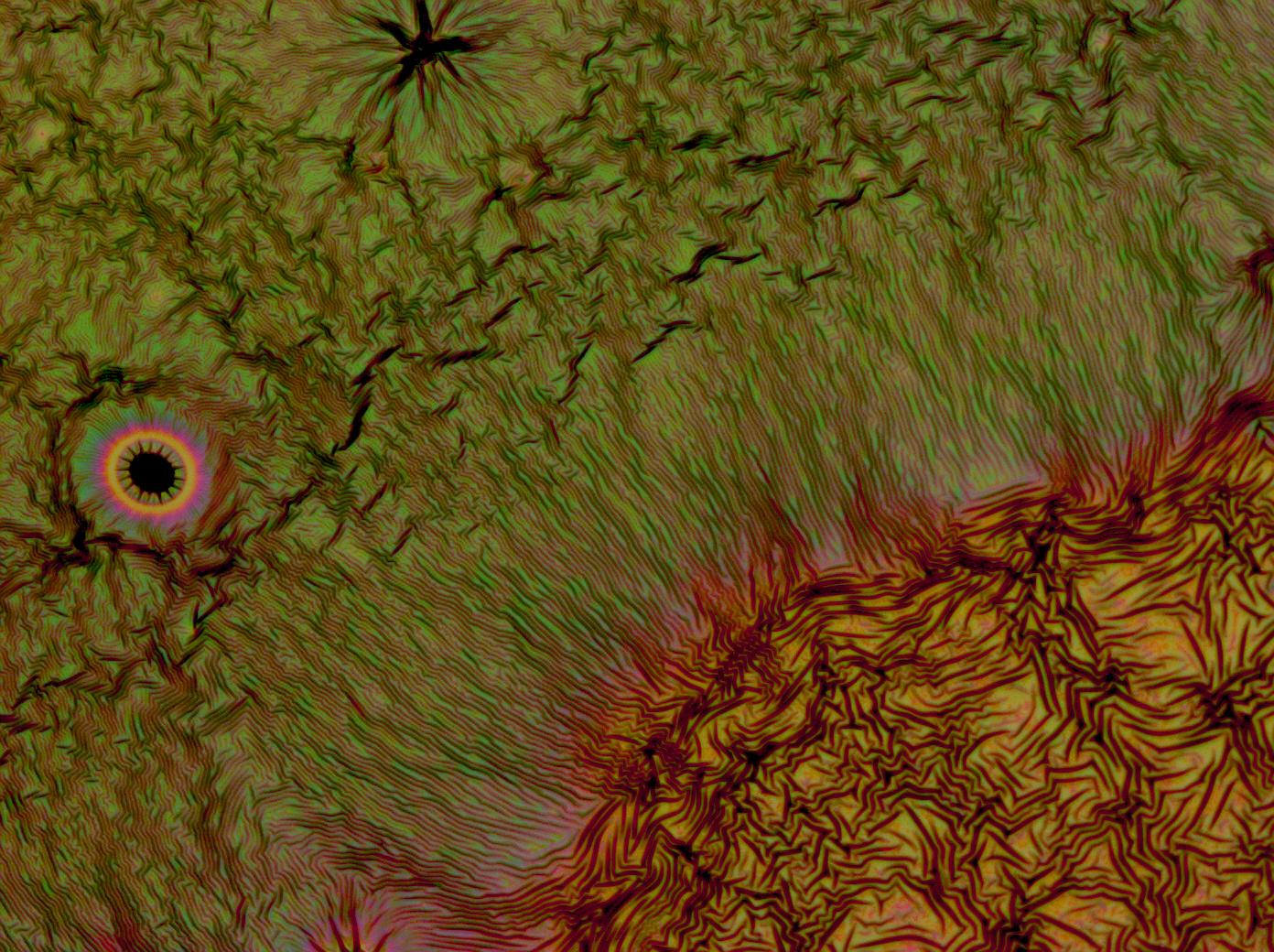
| + | <center> | ||
| + | <gallery caption='A sample of the microscope images taken of multi-layers.' widths=140px> | ||
| + | File:Cam Multilayer drop 1.jpg | ||
| + | File:Cam Crazy multilayer single AP 2k spin2nd.jpg | ||
| + | File:Cam crazy multilayer2.jpg | ||
| + | File:Cam_crazy_multilayer3.jpg | ||
| + | </gallery> | ||
| + | </center> | ||
| + | |||
| + | ==Polyhis-Tagging Reflectin== | ||
| + | |||
| + | The reflectin genes were polyhis-tagged by incorporating the sequence of part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K128005 BBa_K128005] into the primers used to clone the gene. Polyhis tagging was chosen because it allows proteins to be purified with relatively simple apparatus, an affinity column, to which a metal containing resin is added traps the protein when cell lysate is added. | ||
| + | |||
| + | ==Over-Expression== | ||
| + | |||
| + | Reflectin was placed on a high copy plasmid ([http://partsregistry.org/Part:pSB1A2 PSB1A2]) under an arabinose inducible promoter ([http://partsregistry.org/Part:BBa_I0500 pBad]) in order to express reflectin at a high level. As well as a negative control, we used an sfGFP-reflectin fusion as a control for reflecitn production. | ||
| + | |||
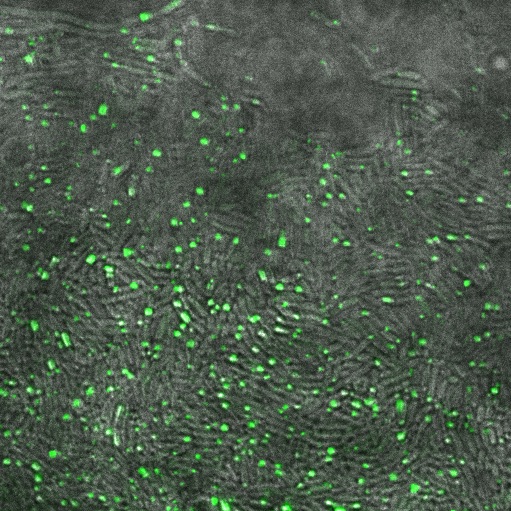
| + | In the non-GFP cells we found that there were brightly lit points at the ends of nearly every cell, which were not present in the negative control. In the GFP fusion cells, these spots glowed strongly, while the rest of the cell was relatively dark. This indicated that reflectin produces inclusion bodies when expressed at high level. | ||
| + | |||
| + | [[File:Cam Reflectin-GFP-inclusionbodies.jpg | thumb| 400px| center| ''E. coli'' transformed with our pBAD-ReflectinA1-GFP construct, induced by adding 1mM arabinose, demonstrating localisation of the protein.]] | ||
| + | |||
| + | ==Extraction & Purification== | ||
| + | We investigated two principal methods for extracting the protein from the transformed <i>E. coli</i> - we lysed the cells and ran the lysate through a polyhis affinity column and we tried a proprietary inclusion body prep. | ||
| + | |||
| + | ===[[Team:Cambridge/Protocols/Protein_Purification | Polyhis-Tag Affinity Column]]=== | ||
| + | The polyhis affinity column allowed us to extract our tagged protein from the lysate without too much difficulty. In order to increase the purity of our sample we tried using an [[Team:Cambridge/Protocols/Acetone_Precipitation_of_Proteins | acetone]] or [[Team:Cambridge/Protocols/Ethanol_Precipitation_of_Protein | ethanol]] precipitation step to remove chemicals retained in the elution buffer and to concentrate the protein for downstream processing. | ||
| + | |||
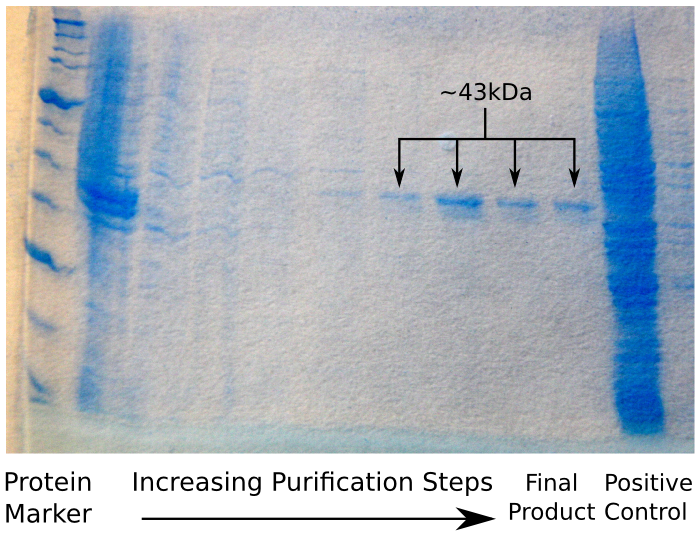
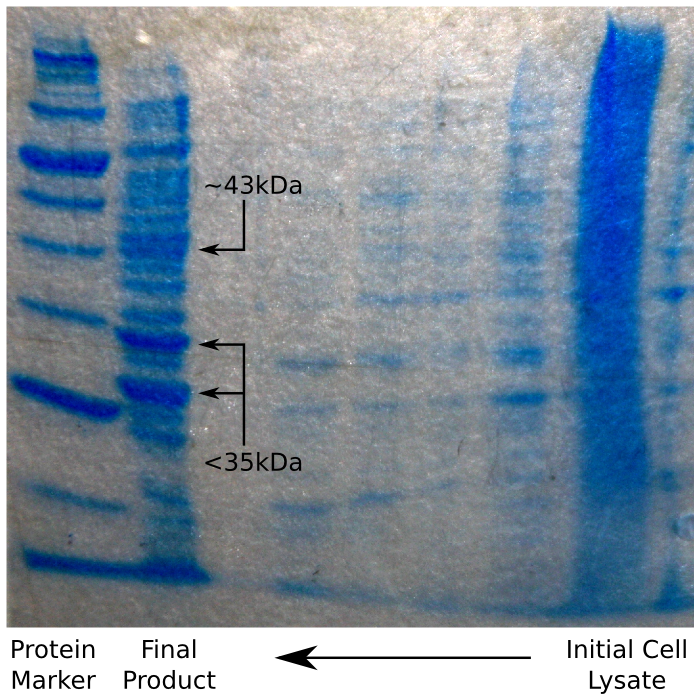
| + | Having done this, we ran our sample on an SDS Page protein gel, to verify that we had in fact purified reflectin. We were expecting to see a thick band of reflectin at around 43 kDa, which – as the image shows – was the case. | ||
| + | |||
| + | [[File:CAM_SDS_POLYHIS.png | center | thumb | 500px | The result of the Polyhis affinity column SDS Page gel]] | ||
| + | |||
| + | ===[[Team:Cambridge/Protocols/Inclusion_Body_Prep | Inclusion Body Prep]]=== | ||
| + | |||
| + | The Norgen Proteospin inclusion body prep kit was experimented with as a commercial alternative to the previous purification method. The kit was simple to use, but required the use of an ultracentrifuge capable of spinning up to 25ml of fluid at 27,000g. It was found that the spinning forces specified in the protocol for these steps weren't capable of moving fluid through the necessary columns and higher forces were needed. | ||
| + | |||
| + | On analysing the end-product of the purification with SDS-PAGE approximately 20 discrete bands were observed from 10-200 kda with 2 particularly bold bands, both of which are too short for reflectin. However, a reasonably thick band of protein was present at 43kDA (the correct length for reflctin). | ||
| + | |||
| + | [[File:CAM_SDS_INCLUSION_BODY.png | center | thumb | 450px | The result of the inclusion body SDS Page gel]] | ||
| + | |||
| + | As the gel shows, the resulting protein is nowhere near as pure as the result of the previous method as the inclusion bodies do not contain only reflectin. However, greater purity might have been achieved by using the result of the inclusion body prep as the first stage of the polyhis affinity protocol, as it would reduce the amount of non-polyhis protein going in to the column. This was not tested in practice, however, as our protein appeared to be pure enough for processing, and we did not have the time to experiment further. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Future Work=== | ||
| + | As well as further improving our thin film's colour, we also wanted to improve their long-term stability, as well as investigate controlling their colour electrically. | ||
{{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} | ||
Latest revision as of 03:10, 22 September 2011
Contents |
Isolating Reflectin and Making Thin Films
Previous in vitro investigations of reflectin showed that it was possible to use a method called flow coating to deposit a thin layer of reflectin onto a silicon substrate which would then demonstrate structural colour.
We wanted to investigate using a different coating method – spin coating – and to try out different methods for protein purification. A polyhis-tagged variant of the protein was overexpressed and purified using a polyhis affinity column. Thin films were made by precipitating with either acetone or ethanol and resuspending in HFIP before either spin coating or flow coating onto a silicon wafer.
We also investigated an inclusion body prep, as we found that the protein formed inclusion bodies upon over-expression. By varying our purification and coating methods, we were able to create vibrant thin films.
Thin Films
In order to demonstrate structural colour, thin films were created by re-suspending the purified protein in HFIP and spin coating or flow coating the re-suspended protein onto a silicon substrate. This gave us brightly coloured thin films which responded to water vapour in the air by changing colour. We performed controls by making HFIP films and films with Bovine Serum Albumin (BSA, a generic protein) neither of which showed any structural colour.
Our initial films were not uniform in colour and would form crystals when allowed to dry out. We improved this by increasing the purity of our protein as well as refining our coating technique.
We first tried to reduce the urea content of our sample, in the belief that this would reduce the tendency to crystallise, however, this did not work as our sample became too dilute. Almost by accident, we tried using a lower volume of protein and found - to our great surprise - that the quality of the thin films produced increased greatly, and the crystallisation was greatly reduced.
The reflectin thin films were dynamic - they responded to water vapour in the air. The video below shows the effect of breathing several times on a thin film - the colour change is quite dramatic.
(See more videos of our thin films [http://www.youtube.com/user/cambridgeigem2011 here]).
Flushed with success, we then pushed on to make multi layers of Reflectin, hoping that this would again increase the brightness and uniformity of the thin films. The beauty of the results was astonishing.
Polyhis-Tagging Reflectin
The reflectin genes were polyhis-tagged by incorporating the sequence of part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K128005 BBa_K128005] into the primers used to clone the gene. Polyhis tagging was chosen because it allows proteins to be purified with relatively simple apparatus, an affinity column, to which a metal containing resin is added traps the protein when cell lysate is added.
Over-Expression
Reflectin was placed on a high copy plasmid ([http://partsregistry.org/Part:pSB1A2 PSB1A2]) under an arabinose inducible promoter ([http://partsregistry.org/Part:BBa_I0500 pBad]) in order to express reflectin at a high level. As well as a negative control, we used an sfGFP-reflectin fusion as a control for reflecitn production.
In the non-GFP cells we found that there were brightly lit points at the ends of nearly every cell, which were not present in the negative control. In the GFP fusion cells, these spots glowed strongly, while the rest of the cell was relatively dark. This indicated that reflectin produces inclusion bodies when expressed at high level.
Extraction & Purification
We investigated two principal methods for extracting the protein from the transformed E. coli - we lysed the cells and ran the lysate through a polyhis affinity column and we tried a proprietary inclusion body prep.
Polyhis-Tag Affinity Column
The polyhis affinity column allowed us to extract our tagged protein from the lysate without too much difficulty. In order to increase the purity of our sample we tried using an acetone or ethanol precipitation step to remove chemicals retained in the elution buffer and to concentrate the protein for downstream processing.
Having done this, we ran our sample on an SDS Page protein gel, to verify that we had in fact purified reflectin. We were expecting to see a thick band of reflectin at around 43 kDa, which – as the image shows – was the case.
Inclusion Body Prep
The Norgen Proteospin inclusion body prep kit was experimented with as a commercial alternative to the previous purification method. The kit was simple to use, but required the use of an ultracentrifuge capable of spinning up to 25ml of fluid at 27,000g. It was found that the spinning forces specified in the protocol for these steps weren't capable of moving fluid through the necessary columns and higher forces were needed.
On analysing the end-product of the purification with SDS-PAGE approximately 20 discrete bands were observed from 10-200 kda with 2 particularly bold bands, both of which are too short for reflectin. However, a reasonably thick band of protein was present at 43kDA (the correct length for reflctin).
As the gel shows, the resulting protein is nowhere near as pure as the result of the previous method as the inclusion bodies do not contain only reflectin. However, greater purity might have been achieved by using the result of the inclusion body prep as the first stage of the polyhis affinity protocol, as it would reduce the amount of non-polyhis protein going in to the column. This was not tested in practice, however, as our protein appeared to be pure enough for processing, and we did not have the time to experiment further.
Future Work
As well as further improving our thin film's colour, we also wanted to improve their long-term stability, as well as investigate controlling their colour electrically.
 "
"