Team:Brown-Stanford/REGObricks/Transforming
From 2011.igem.org
(→Troubleshooting) |
(→Troubleshooting) |
||
| (2 intermediate revisions not shown) | |||
| Line 88: | Line 88: | ||
We speculated that repeated centrifuge at high speeds was too traumatic for the digested protoplast cells, and the survival rate following the transformation was too low to produce colony forming units. | We speculated that repeated centrifuge at high speeds was too traumatic for the digested protoplast cells, and the survival rate following the transformation was too low to produce colony forming units. | ||
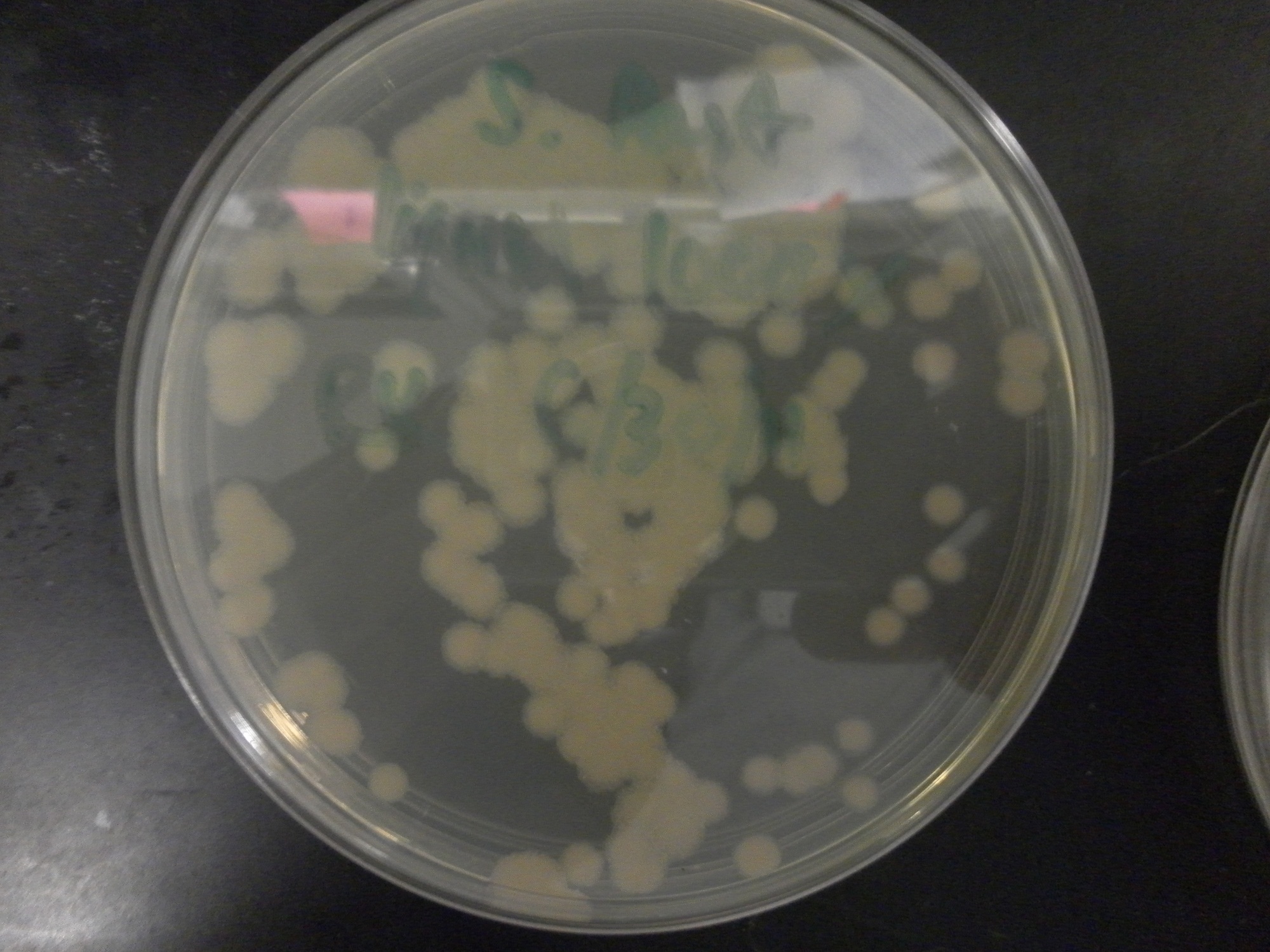
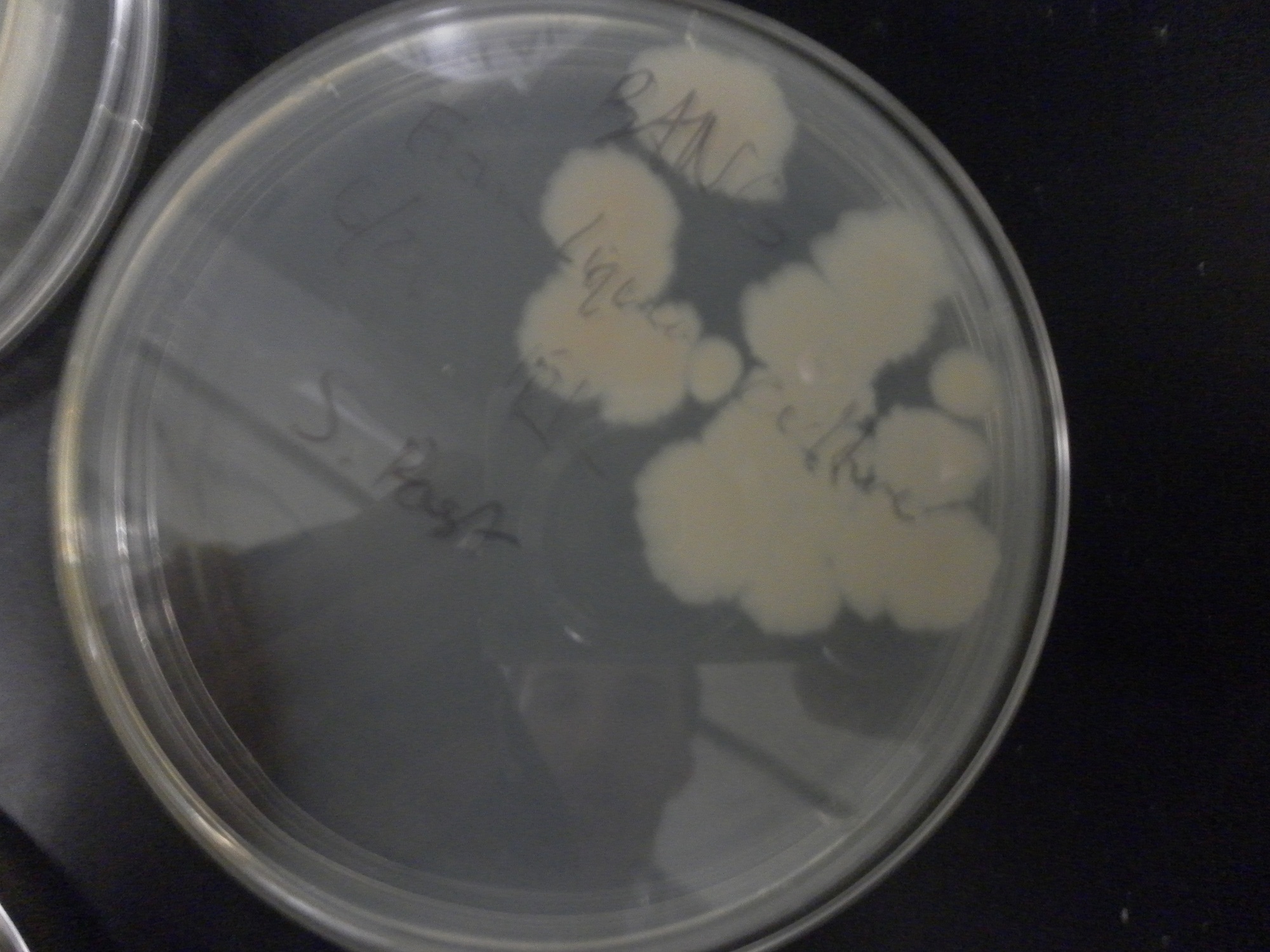
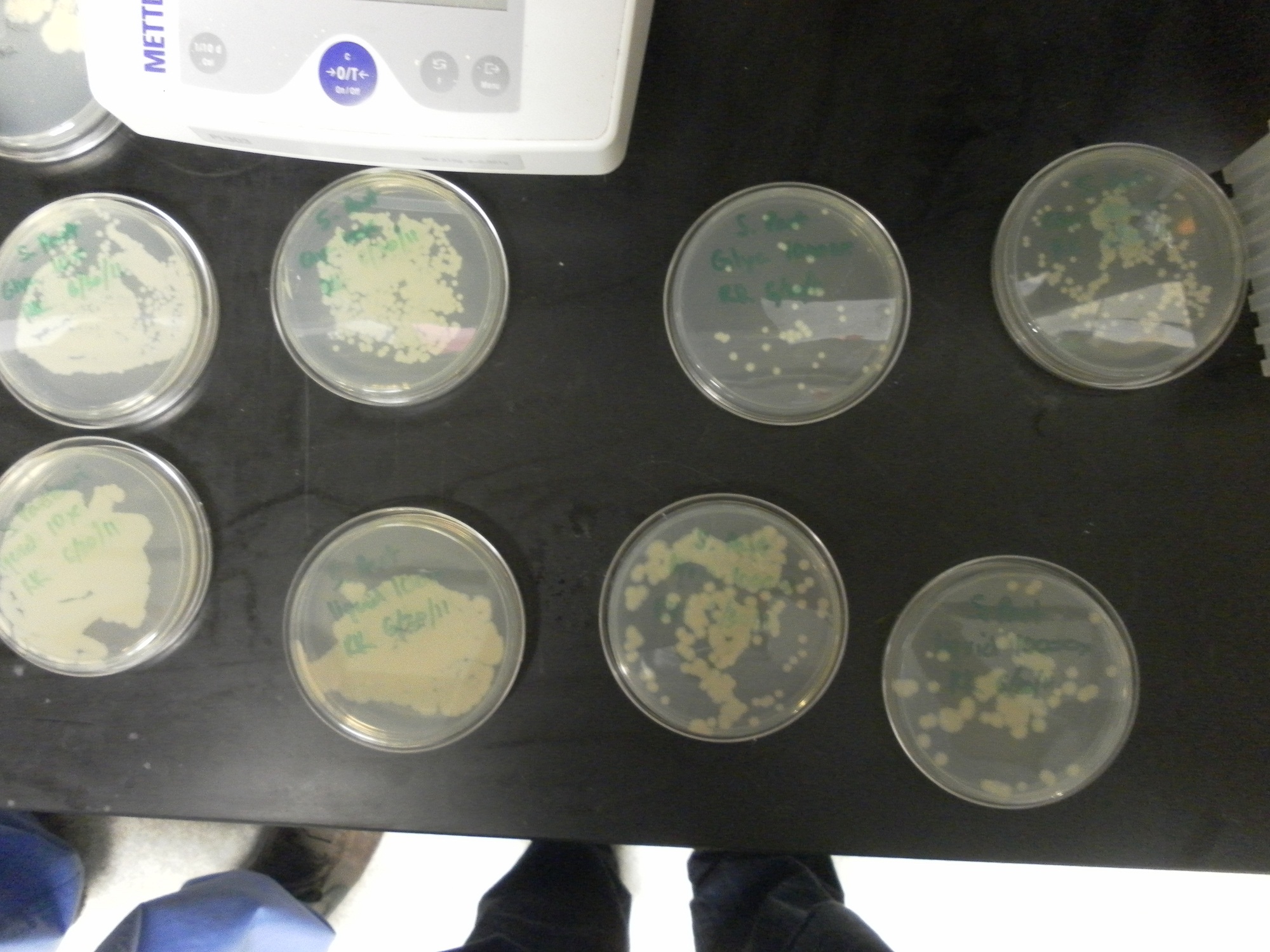
| - | Additional difficulties also arose in the recovery phase of the protoplast transformation, as the lack of antibiotic selection sometimes compromised portions of our rich recovery plates. (The recovery plates contained over 340 g of sucrose/mL).<center> [[File:Brown-Stanford-Contaminatedcells.JPG|300px|none|thumb|Suspected Contamination of ''S. pasteurii'' Transformation ]] [[File:Brown-Stanford-Contaminatedcells2.JPG|300px|none|thumb|Suspected Contamination of ''S. pasteurii'' Transformation ]] [[File:Brown-Stanford-Contaminatedcells3.JPG|300px|none|thumb|Suspected Contamination of ''S. pasteurii'' Transformation ]] | + | Additional difficulties also arose in the recovery phase of the protoplast transformation, as the lack of antibiotic selection sometimes compromised portions of our rich recovery plates. (The recovery plates contained over 340 g of sucrose/mL). <center> [[File:Brown-Stanford-Contaminatedcells.JPG|300px|none|thumb|Suspected Contamination of ''S. pasteurii'' Transformation ]] [[File:Brown-Stanford-Contaminatedcells2.JPG|300px|none|thumb|Suspected Contamination of ''S. pasteurii'' Transformation ]] [[File:Brown-Stanford-Contaminatedcells3.JPG|300px|none|thumb|Suspected Contamination of ''S. pasteurii'' Transformation ]]</center> |
=== '''Conclusions''' === | === '''Conclusions''' === | ||
Latest revision as of 04:08, 29 September 2011
Transformation
Due Respect
Those familiar with the iGEM competition might remember a previous predecessor who also focused on the microbial carbonate precipitation - Bacilla Filla, Team Newcastle 2010’s project investigated the potential of Bacillus subtilis to remediate fractures in concrete architecture. However, due to the finicky nature of urease, they decided to focus on upregulating urea production inside the cell for their project.
“Previous experiments involving up-regulating ureA, ureB and ureC in B. subtilis have not lead to an increase in urease production. This could be due to yet unidentified genes that are involved in the process. Therefore, we looked for another strategy.” – Newcastle 2010
Since the last Jamboree, the study of microbial precipitation has risen in prominence beyond just remediation. As we suggest in our project, it is instead being championed as a new way to aggregate building materials entirely. This expansion is related to the robust urease function present in the new organism of study: S. pasteurii.
Motivation
Though B. subitilis has more documentation of successful transformations, we decided to embark on a path less traveled by, to attempt to transform S. pasteurii. Characterization from recent research has shown numerous attractive features intrinsic to the S. pasteurii bacterium, including
- Robust function of its urease activity, even in 100% salt water 1.
- Its ability to utilize waste products and proliferate from cheap sources of nutrition (ie. Corn steep liquor, vegemite, etc)2.
- The success of mutagenic experiments to increase urease function 3.
Correspondingly, the motivation to investigate the transformation of S. pasteurii comes from several key benefits that would be accomplished if we were successful:
- The ability to confer antibiotic resistance to the cell culture, opening the door to
- More rigorous genetic controls cell culture fidelity**
- Allowing continued selection in directed evolution experiments
- The ability to introduce traits that will strengthen the bacterium profile for extraterrestrial space travel
With a successful path to genetic recombination, we could add this organism to our list of powerful chassis’s, for use both terrestrial and cosmic use.
**During the course of our cell culture of ''S. pasteurii'', we frequently encountered contamination of our media-rich culture, despite our best attempts at sterilization techniques under the UV hood.
The risk involved in this project selection was the existence of good reasons for the lack of published articles on S. pasteurii transformation. But in the words of Microsoft’s blue monster, we decided that we were going to either go big or go home.
Experiment Design
Through all of the research we scoured, we found only one mention of the genetic transformation of S. pasteurii, from a symposium done by a then-undergraduate student in Arizona State University. In it she documented a seemingly successful transformation of S. pasteurii using protoplast transformation, giving us hope of its feasibility. Through fortuitous email correspondence, we were able establish contact with Alicia, who had had went on to become a practicing attorney. She kindly faxed us her old lab notebook sheets. We are still in awe of her organization skills.
Several methods of transformation were available to choose from:
- Soil Transformation- relying on the natural competency of the bacteria to transfer genetic information
- Protoplast Transformation- Digestion of the cell wall with lysozymes, followed by induction of DNA uptake by PEG 8000.
- Electroporation- research suggests limited success with gram positive cells
- Protoplast and Electroporation- a combination of two techniques, use of lysozymes to digest cell wall and electroporation to create entrances for DNA
Because of S. pasteurii’s thick cell wall, the protoplast transformation seemed the most logical to succeed.
Transformation
S. pasteurii cells were obtained via ATCC 11859 stock. The transforming DNA was obtained from Dr. Yun-Peng Chao, Professor of Chemical Engineering at Feng Chia University in Taiwan. Plasmid pUB110 (not to be confused with pBU11 , was a shuttle vector between E. coli and B. subtilis, containing an Ampicillin cassette for selection of the former, and a Kanamycin cassette for the latter.
pUB110 was selected following the protocols suggested by Alicia’s notes, reasoning along the lines that the homology between the two bacteria (S. pasteurii was previously known as Bacillus pasteurii before a recategorization) would be sufficient to allow expression of the Kanamycin resistance cassette inside S. pasteurii.
Newcastle iGEM’s 2009 Sporulation Part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K174011 BBa_K174011] was also received via request to the Registry of Standardized Parts.
Protocols for the transformation can be found here.
Lysozyme from chicken egg white (L3790 - 10 mg/mL) was obtained from Sigma-Aldrich.
Troubleshooting
Many attempts at protoplast transformation were carried out, but the results were unsuccessful: there was either no growth on the Kanamycin resistance plates, or there was excessive growth, with distinctly different colony features and tested negative on urea-phenol red test plates. This suggested that there was contamination due to 1) improperly prepared kanamycin plates or 2) kanamyci1n resistant strains colonizing the extremely rich recovery plates.
Troubleshooting suggested that the capacity of the protoplasts to regenerate their cell wall was a potential limiting factor. Viewing of the lysozyme digestion showed nearly complete digestion of the cell membranes (before and after). Procedures for collecting the protoplasts involved several centrifugation step at an unspecified “maximum speed” according to the protocol, though there was difficulty in collecting cell pellets at speeds below 8000 g, especially after the addition of viscous PEG 8000.
We speculated that repeated centrifuge at high speeds was too traumatic for the digested protoplast cells, and the survival rate following the transformation was too low to produce colony forming units.
Additional difficulties also arose in the recovery phase of the protoplast transformation, as the lack of antibiotic selection sometimes compromised portions of our rich recovery plates. (The recovery plates contained over 340 g of sucrose/mL).Conclusions
In retrospect, we were a little too ambitious in choosing the scale of our projects. The elucidation of successful transformation protocols required more time than we had available, and at the time of this wikifreeze, we are still trying different concentrations of PEG, and lysozyme digestion on S. pasteurii.
Our plan to characterize the function of 2010 Newcastle’s sporulation part is also currently unrealized, due to the difficulties in transforming S. pasteurii. Nevertheless, we believe that though it has not been realized for the cut-off date of this jamboree, sophisticated investigation of S. pasteurii as a genetic chassis is still a worthwhile endeavor. Particularly as the industrial commercialization of biocementation reaches fruition, the understanding and ability to regulate the genetic activity of urease in S. pasteurii will be an essential breakthrough optimizing and fine-tuning the biostructures we create.
References
1 Mortensen B.M. et al. Effects of environmental factors on microbial induced
calcium carbonate precipitation
Journal of Applied Microbiology 111, 338–349]]
2 Reddy, M. S. et al. Biocalcification by Sporosarcinapasteurii using corn steep liquoras nutrient source. . J Ind Microbiol Biotechnol June 2010, 6(3): 170-174.]
3 Achal, V. et al. Strain improvement of Sporosarcina pasteurii
for enhanced urease and calcite production. J Ind Microbiol Biotechnol (2009) 36:981–988.]
 "
"