Team:Brown-Stanford
From 2011.igem.org
(→APPLYING FOR STANFORD iGEM 2012) |
|||
| (19 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
{{:Team:Brown-Stanford/Templates/Twitter}} | {{:Team:Brown-Stanford/Templates/Twitter}} | ||
| - | = ''' | + | = '''APPLY TO JOIN US IN 2012''' = |
| + | The Brown and Stanford iGEM teams are planning to continue our transcontinental collaboration in the upcoming year. If you are a student at either of our universities and are interested in participating, please follow the recruitment instructions for your location! | ||
| + | Still confused about exactly what Synthetic Biology is? Not to worry, here's an awesome video explaining it! http://www.youtube.com/watch?v=rD5uNAMbDaQ&feature=youtu.be | ||
| + | |||
| + | |||
| + | == '''For Stanford students''' == | ||
Please fill out our interest form at https://docs.google.com/spreadsheet/viewform?formkey=dEQyaWdReTN5S2pCcHRVY29hQXh4cHc6MQ. | Please fill out our interest form at https://docs.google.com/spreadsheet/viewform?formkey=dEQyaWdReTN5S2pCcHRVY29hQXh4cHc6MQ. | ||
| - | We will be sending further information to | + | Applications will be due Sunday, February 5th at 11:59 PM. Please fill out the application form [http://dl.dropbox.com/u/10558719/2012_iGEM_Application.doc here], and also attach your resume and (unofficial) transcript and email them to stanfordigem@gmail.com. |
| + | |||
| + | We'll be holding some lab technique boot camps and brainstorming/planning sessions in the spring if you make the team, so expect that. If you're chosen, you should also take Dr. Drew Endy's BioE 44 Synthetic Biology Lab in the spring, which is, by the way, an awesome class. | ||
| + | |||
| + | We will be sending further information to the list above! | ||
Thanks! | Thanks! | ||
| - | = ''' | + | == '''For Brown students''' == |
(12/5/11) | (12/5/11) | ||
Brown iGEM is looking for undergrad team members for this upcoming year! | Brown iGEM is looking for undergrad team members for this upcoming year! | ||
| Line 33: | Line 42: | ||
<html> | <html> | ||
| - | <span style="font-size: 20px;"></html> | + | <span style="font-size: 20px;"></html>The electronic application at Brown is [http://www.brownigem.com/2012_iGEM_Application.doc available now]. Deadline is 11:59pm on 1/31! |
| + | |||
| + | |||
| + | Keep up with the 2012 application process on our [https://www.facebook.com/events/146221565480758/ facebook page]! | ||
<html></span></html> | <html></span></html> | ||
| Line 97: | Line 109: | ||
=== '''Latest Advancements on Projects''' === | === '''Latest Advancements on Projects''' === | ||
| - | ===== ''' | + | ===== '''Anabaena Transformation and Successful Cell-Type Specific Expression Control''' ===== |
| + | We successfully transformed our cyanobacterial chassis, Anabaena PCC 7120, with our designed PowerCell genetic construct. Unlike more common cyanobacteria species such as Synechocystis PCC 6803 or Synechococcus elongatus PCC 7942, Anabaena is not naturally competent and does not easily uptake foreign DNA. We performed a triparental conjugation in order to bypass the natural barriers to transformation. | ||
| - | + | To demonstrate that expression of our construct is properly co-localized to vegetative cells, we grew cultures of Anabaena in nitrate-free BG11 media to promote heterocyst formation. We demonstrated the successful control of our pSac promoter over our sugar secretion construct through GFP expression. The sugar secretion construct is only expressed in actively photosynthesizing cells. In the first picture, we observe areas of diminished GFP fluorescence at regular intervals (approximately 1 in 10 cells) along our transformed Anabaena filament (the background fluorescence is likely due to chlorophyll). Since our GFP reporter should only express under the pSac promoter and only in non-heterocyst cells, we would expect the spacing of dark spots to match the spacing of heterocysts identified through alcian blue staining. Alcian blue is a dye that selectively binds to polysaccharide chains on the surface of heterocyst cells. As you can see by comparing the two images, our results appear to match up! | |
| - | + | ||
| - | + | ||
| - | + | Work to characterize the performance of our cscB sucrose secretion construct is underway. We will assay sucrose levels produced by PowerCell. If they are high enough, we will attempt to grow E. coli W. containing arbitrary biobricks from this nutrient source, in an attempt to show PowerCell can power biological tools. | |
| - | < | + | <center> |
| + | <html><object width="400" height="300"> <param name="flashvars" value="offsite=true&lang=en-us&page_show_url=%2Fphotos%2Fbrownstanfordigem%2Fsets%2F72157628308638459%2Fshow%2F&page_show_back_url=%2Fphotos%2Fbrownstanfordigem%2Fsets%2F72157628308638459%2F&set_id=72157628308638459&jump_to="></param> <param name="movie" value="http://farm8.staticflickr.com/7030/6467315137_890d8a769b_o.jpg"></param> <param name="allowFullScreen" value="true"></param><embed type="application/x-shockwave-flash" src="http://farm8.staticflickr.com/7030/6467315137_890d8a769b_o.jpg" allowFullScreen="true" flashvars="offsite=true&lang=en-us&page_show_url=%2Fphotos%2Fbrownstanfordigem%2Fsets%2F72157628308638459%2Fshow%2F&page_show_back_url=%2Fphotos%2Fbrownstanfordigem%2Fsets%2F72157628308638459%2F&set_id=72157628308638459&jump_to=" width="400" height="300"></embed></object></html> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<div style="width: 35%;"> | <div style="width: 35%;"> | ||
| - | Transformed Anabaena | + | Transformed Anabaena showing cell-type specific GFP fluorescence under the control of the pSac promoter. GFP expression is on in vegetative cells, off in heterocysts. Some background fluorescence from chlorophyll is visible. |
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | ===== | + | <html><object width="400" height="300"> <param name="flashvars" value="offsite=true&lang=en-us&page_show_url=%2Fphotos%2Fbrownstanfordigem%2Fsets%2F72157628308638459%2Fshow%2F&page_show_back_url=%2Fphotos%2Fbrownstanfordigem%2Fsets%2F72157628308638459%2F&set_id=72157628308638459&jump_to="></param> <param name="movie" value="http://farm8.staticflickr.com/7152/6467315073_227d36b07a_o.jpg"></param> <param name="allowFullScreen" value="true"></param><embed type="application/x-shockwave-flash" src="http://farm8.staticflickr.com/7152/6467315073_227d36b07a_o.jpg" allowFullScreen="true" flashvars="offsite=true&lang=en-us&page_show_url=%2Fphotos%2Fbrownstanfordigem%2Fsets%2F72157628308638459%2Fshow%2F&page_show_back_url=%2Fphotos%2Fbrownstanfordigem%2Fsets%2F72157628308638459%2F&set_id=72157628308638459&jump_to=" width="400" height="300"></embed></object></html> |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<div style="width: 35%;"> | <div style="width: 35%;"> | ||
| - | Transformed Anabaena | + | Transformed Anabaena with heterocysts stained by alcian blue. Heterocyst spacing corresponds to the "off" cells from the GFP image, implying that the sucrose secretion construct in not active in heterocysts, as desired. |
</div> | </div> | ||
| - | |||
| - | |||
</center> | </center> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
===== '''Biobricked the urease operon''' ===== | ===== '''Biobricked the urease operon''' ===== | ||
Latest revision as of 19:40, 11 January 2012
APPLY TO JOIN US IN 2012
The Brown and Stanford iGEM teams are planning to continue our transcontinental collaboration in the upcoming year. If you are a student at either of our universities and are interested in participating, please follow the recruitment instructions for your location!
Still confused about exactly what Synthetic Biology is? Not to worry, here's an awesome video explaining it! http://www.youtube.com/watch?v=rD5uNAMbDaQ&feature=youtu.be
For Stanford students
Please fill out our interest form at https://docs.google.com/spreadsheet/viewform?formkey=dEQyaWdReTN5S2pCcHRVY29hQXh4cHc6MQ.
Applications will be due Sunday, February 5th at 11:59 PM. Please fill out the application form [http://dl.dropbox.com/u/10558719/2012_iGEM_Application.doc here], and also attach your resume and (unofficial) transcript and email them to stanfordigem@gmail.com.
We'll be holding some lab technique boot camps and brainstorming/planning sessions in the spring if you make the team, so expect that. If you're chosen, you should also take Dr. Drew Endy's BioE 44 Synthetic Biology Lab in the spring, which is, by the way, an awesome class.
We will be sending further information to the list above!
Thanks!
For Brown students
(12/5/11) Brown iGEM is looking for undergrad team members for this upcoming year!
Have you ever wanted to engineer living organisms to create novel functions and help transform the world?
Spend a summer working at the Synthetic Biology Initiative at NASA Ames Research Center, and represent Brown at the premiere synthetic biology competition in the world! Opportunities exist for all kinds of microbiology and molecular biology work, computational biology, mathematical modeling, engineering, and much more. Develop incredible research and lab skills as you manage your own project!
Students of all academic backgrounds and experience levels welcome (this includes Class of '12)
We are hosting two INFO SESSIONS this week:
TIME: 6-7pm on Thursday 12/8, Friday 12/9
LOCATION: Barus and Holly 190
-come hear more about iGEM, synthetic biology, and what the award-winning Brown iGEM team does; dinner will be provided!
If you can't make it but are still interested, please let us know! Other questions or concerns are welcome as well. Contact us at igem@brown.edu
The electronic application at Brown is [http://www.brownigem.com/2012_iGEM_Application.doc available now]. Deadline is 11:59pm on 1/31!
Keep up with the 2012 application process on our facebook page!
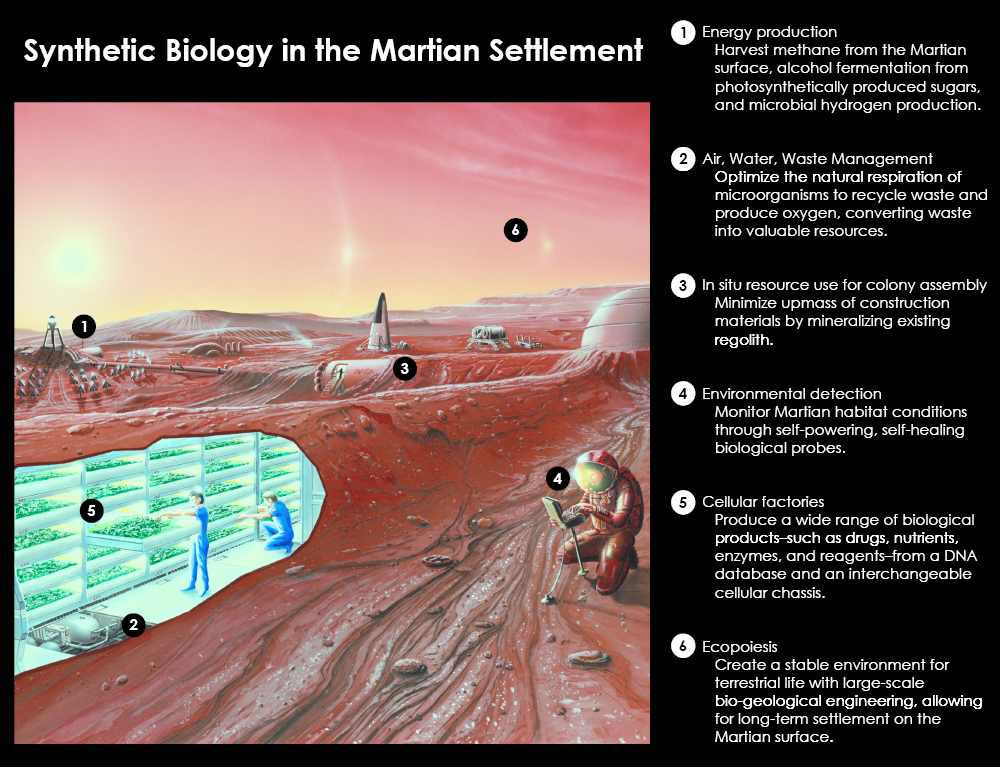
Mars BioTools: Synthetic Biology for Space Exploration
One of the major challenges of space exploration is the limited payload mass that can be launched on a rocket and the difficulty of resupply mid-mission. Any long term settlement will require more resources than astronauts can initially bring with them. Synthetic Biology has the potential to revolutionize space exploration and settlement. Biological tools have a major advantage over classical tools: the ability to self-replicate and regenerate.
The emerging field of Synthetic Biology will allow us to engineer microbial factories that will largely circumvent the limiting payload factors. These cellular factories will generate fuel, food, medicines and building materials for settlers, but will consist of engineered cells that could be stored in a single test tube, and regrown to production scale on-site, as needed.
The Brown-Stanford iGEM team is excited to work on three different projects, under the common theme of developing Synthetic Biology applications for space. In addition, as a response to the ethical and philosophical considerations, we interviewed prominent researchers and leaders in the field. Click on the astronaut in the corner to view our series.
 REGObricks
REGObricks
Any extra-terrestrial settlement will require a habitat to keep its occupants alive, but transport of such a habitat will require a huge amount of payload space. RegoBricks uses bacteria to cement Martian and Lunar regolith (soil) simulant into durable building blocks, similar to concrete bricks, allowing settlements to use in situ resources to build structures.
 FRETcetera
FRETcetera
Biological interfaces cannot pass information to their users as fast as electronic ones can. FRETcetera develops a novel method of fast-acting biological reporting with changes in cell fluorescence. Bacteria could be used to detect toxic chemicals in the environment, for example, or inform astronauts that their microbial tools are unhealthy.
 PowerCell
PowerCell
All biological tools need energy to run. PowerCell develops a universal energy source. Engineered photosynthetic bacteria generate carbon and nitrogenous nutrients from sunlight and air and secrete them to power biological tools. These tools will transform the raw materials into fuel, building materials, food, drugs, and other products useful to settlers.
Summary
News!
iGEM Awards
We did very well at iGEM this year!
-At the Americas Regional Jamboree, we finished Top 4 and won Best Presentation.
-At the World Jamboree, we finished in the Sweet 16, and won Best New Application.
We are extremely grateful to all those who helped us along the way, and our mentors. Thank you so much!
Project Summary
Here's a minute long teaser of our project!
Latest Advancements on Projects
Anabaena Transformation and Successful Cell-Type Specific Expression Control
We successfully transformed our cyanobacterial chassis, Anabaena PCC 7120, with our designed PowerCell genetic construct. Unlike more common cyanobacteria species such as Synechocystis PCC 6803 or Synechococcus elongatus PCC 7942, Anabaena is not naturally competent and does not easily uptake foreign DNA. We performed a triparental conjugation in order to bypass the natural barriers to transformation.
To demonstrate that expression of our construct is properly co-localized to vegetative cells, we grew cultures of Anabaena in nitrate-free BG11 media to promote heterocyst formation. We demonstrated the successful control of our pSac promoter over our sugar secretion construct through GFP expression. The sugar secretion construct is only expressed in actively photosynthesizing cells. In the first picture, we observe areas of diminished GFP fluorescence at regular intervals (approximately 1 in 10 cells) along our transformed Anabaena filament (the background fluorescence is likely due to chlorophyll). Since our GFP reporter should only express under the pSac promoter and only in non-heterocyst cells, we would expect the spacing of dark spots to match the spacing of heterocysts identified through alcian blue staining. Alcian blue is a dye that selectively binds to polysaccharide chains on the surface of heterocyst cells. As you can see by comparing the two images, our results appear to match up!
Work to characterize the performance of our cscB sucrose secretion construct is underway. We will assay sucrose levels produced by PowerCell. If they are high enough, we will attempt to grow E. coli W. containing arbitrary biobricks from this nutrient source, in an attempt to show PowerCell can power biological tools.
Transformed Anabaena showing cell-type specific GFP fluorescence under the control of the pSac promoter. GFP expression is on in vegetative cells, off in heterocysts. Some background fluorescence from chlorophyll is visible.
Transformed Anabaena with heterocysts stained by alcian blue. Heterocyst spacing corresponds to the "off" cells from the GFP image, implying that the sucrose secretion construct in not active in heterocysts, as desired.
Biobricked the urease operon
We successfully PCR cloned a 10.7 kb functional urease cassette from the plasmid pBU11, whose genetic sequence has hitherto never been known before. We ligated it onto a backbone, and successfully made a standard BioBrick, characterizing its function and demonstrating active urease activity. Urease is the catalyst behind the phenomenon known as biocement, which is able to fuse sand into bricks, using nothing more than calcium ions and urea. More about urease.
Biobrick part [http://partsregistry.org/Part:BBa_K656013 BBa_K656013] has been submitted to the Registry of Standardized Parts. The sequence of this cassette is unavailable because it currently does not exist in any known registry. At the time of this writing, it is currently being sequenced in the lab of Dr. Chris Mason at Weill Cornell Medical College. As soon as the sequence is available, we will supplement the directory listing.
Stratospheric Balloon Flight
We launched samples of our bacteria to the far edge of space (80,000 ft-110,000 ft) to test for their ability to survive simulated Martian environmental conditions. The pressures, temperatures, and radiation conditions at these altitudes are similar to the conditions on the Martian surface. Read about our escapades here
Advancements in Human Practices
Feature on Synthetic Biology and Space
We talked with key figures in science and science policy about the implications of human expansion into space, extraterrestrial life and synthetic biology. You can look at our interviews and findings here or by clicking on the astronaut on the upper left!
Education and Publicity

Editor's Choice! x2
|

Lunar Science Poster. Click to see full version.
|
We educated and publicized the power of synthetic biology in a whole suite of venues and mediums, including
- A crowd of 600+ over two days at Maker Faire NYC, resulting in two Editor’s Choice blue ribbons!
- A filming opportunity with the BBC Horizon, to be featured in a documentary on NASA’s Initiative in Synthetic Biology
- Verbal presentations to NASA Administrator Charlie Bolden, NASA Ames Research Center Director Pete Worden, and writer and visionary Stewart Brand
- A poster presentation at NASA’s Lunar Science Forum
- A heart-pounding poem at the first-ever SB5.0 Synbio Slam
Outreach and Collaboration

We spearheaded the largest collaborative effort in iGEM history to change the way we do outreach, creating [http://openwetware.org/wiki/IGEM_Outreach CommunityBricks], a database of valuable teaching plans, presentations, community resources and projects for young and old alike.
Alumni and Community-Building
We built the [http://alumni.brownigem.com first iGEM Alumni Network,] to provide iGEM'ers a growing collection of professional and social resources and keep the spirit of iGEM alive post-Jamborees.
 "
"