Team:BU Wellesley Software/Trumpet
From 2011.igem.org
Craiglaboda (Talk | contribs) |
|||
| (38 intermediate revisions not shown) | |||
| Line 23: | Line 23: | ||
/*actual content styles*/ | /*actual content styles*/ | ||
| - | body {width: | + | body {width: 800px; margin:auto;} |
#bu-wellesley_wiki_content {height:auto; line-height:100%;} | #bu-wellesley_wiki_content {height:auto; line-height:100%;} | ||
| - | #bu-wellesley_wiki_content a {color:#69d01d;} | + | /*#bu-wellesley_wiki_content a {color:#69d01d;}*/ |
| - | #bu-wellesley_wiki_content a:hover {text-decoration:none; color:# | + | #bu-wellesley_wiki_content a:hover {text-decoration:none; color:#3d3f3c;} |
| - | + | .navbar li {color: #ffffff;} | |
| - | + | .navbar li a {color: #ffffff;} | |
| + | .navbar li a:hover {background:#69d01d; color: #ffffff;} | ||
| + | /*only use for current page content header (i.e. Team, G-nomeSurferPro, etc)*/ | ||
| + | H6 { | ||
| + | font-family: Helvetica; | ||
| + | text-transform: uppercase; | ||
| + | text-decoration: none; | ||
| + | text-align: left; | ||
| + | color: #3d3f3c; | ||
| + | font-size: 32pt; | ||
| + | } | ||
| + | </style> | ||
| + | <link rel="stylesheet" type="text/css" href="http://cs.wellesley.edu/~hcilab/iGEM_wiki/css/Team.css"> | ||
</head> | </head> | ||
<body class="basiclayout"> | <body class="basiclayout"> | ||
| - | |||
<div id="bu-wellesley_wiki_content"> | <div id="bu-wellesley_wiki_content"> | ||
| + | |||
<p style="text-align:center;"><a href="https://2011.igem.org/Team:BU_Wellesley_Software"><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/banner.png" width="800px"></a></p> | <p style="text-align:center;"><a href="https://2011.igem.org/Team:BU_Wellesley_Software"><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/banner.png" width="800px"></a></p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <ul id="nav"> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Team">Team</a></li> | ||
| + | <li><a href="#">Project</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Project_Overview">Overview</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Clotho">Clotho</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/G-nomeSurferPro">G-nome Surfer Pro</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/OptimusPrimer">Optimus Primer</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Trumpet">Trumpet</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/eLabNotebook">eLabNotebook</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Wet_Lab">Wet Lab</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Downloads_and_Tutorials">Downloads and Tutorials</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#">Process</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Methodology">Methodology</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Safety">Safety</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Notebook">Notebook</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Outreach">Outreach</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Tips">Tips and Tricks</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Gold">Medal Fulfillment</a></li> | ||
| + | <li><a href="#">Additional Info</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Acknowledgement">Acknowledgement</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Social">Fun</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| - | < | + | <br> |
| - | + | <h6>Trumpet</h6> | |
| - | < | + | <div id="tracking_nav"> |
| + | JUMP TO...<br> | ||
| + | <a href="#bu-wellesley_wiki_content">Top</a><br> | ||
| + | <a href="#tooloverview">Tool Overview</a><br> | ||
| + | <a href="#method">Method</a><br> | ||
| + | <a href="#example">Design Example</a><br> | ||
| + | <a href="#download">Tutorial and Download</a><br> | ||
| + | <a href="#tutorial">Tabletop Demo</a><br> | ||
| + | <a href="#results">Results</a><br> | ||
| + | <a href="#futurework">Future Work</a> | ||
| + | </div> | ||
| - | + | <div id ="tooloverview"> | |
| - | + | <h1>Tool Overview</h1> | |
| - | < | + | <p> |
| - | + | We are developing a synthetic testbed design for a DNA molecule, | |
| - | Trumpet | + | which we call <em>Trumpet</em>. A trumpet testbed comprises a library of genes and promoters, which can |
| - | + | be configured into any desired permutation or combination by treating the DNA with a sequence of | |
| - | + | enzymes called recombinases. Our design algorithm takes a set of DNA parts, and produces a testbed | |
| - | + | design and enzyme key dictionary, such that the testbed can be configured into any sequence of | |
| - | + | promoters and genes by treating it with the corresponding enzyme key. This allows us to | |
| - | + | rewire the genes and switches and study all their combinations. | |
| - | + | <p> | |
| - | + | <img style="float:right; width:270px; height:250px padding:10px" src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/Trumpet/TrumpetLogo.png"/> | |
| - | <img style="float: | + | The goal of Trumpet is to aid the user in designing fully reconfigurable genetic circuit plasmids. Imagine that as a researcher, you have a particular set of genes, and you are interested in how the expressions of these genes interact with one another. Conventionally, in order to do this, you must tediously construct many different plasmids, each with the correct combinations and permutations of these genes, and study the expressions of these plasmids one at a time. |
| + | <p> | ||
| + | However, what if you could make a single plasmid containing the genes of interest, and use enzymatic reactions to reconfigure the genes into the different permutations and combinations? Not only would this save a great deal of time in the building phase, but it would open the door to a world of completely reconfigurable genetic circuits based on user input via recombinase embedded suicide vectors. | ||
| + | Trumpet is our first attempt at this dream. We have developed a software tool that takes in a set of genes along with the user’s desired set of permutations/combinations, and returns to the user an intelligently designed plasmid which can be reconfigured into the permutations/combinations specified by the user. Along with the plasmid design, Trumpet outputs a set of “keys” for each permutation/combination. These keys indicate the ordered set of recombinase inputs which must be introduced by the user in order to reconfigure the plasmid into the proper state. By developing Trumpet as a Clotho application, the user can quickly save this design to their database of parts. Then, they can use Puppetshow, another one of our applications, to quickly and efficiently build the plasmid using a liquid handling robot. | ||
| + | <p> | ||
| + | <div id="method"> | ||
| + | <h1>Method</h1> | ||
| + | <p> | ||
| + | For this project, we focused on the use of invertases as our recombinase inputs. By orienting two identical copies of an att site in opposite directions, the corresponding invertase which recognizes these att sites will effectively flip the entire duplex region located between the sites. Using this property, we developed two separate algorithms which place at sites intelligently between and around the genes of interest, such that all of the desired combinations and permutations can be achieved through these enzymatic inversions. | ||
| + | <p> | ||
| + | The output of our Pancake and LinkSort translations is a large composite part-containing set of genes with att sites properly inserted such that all permutations and combinations of the genes can be achieved given a particular set of invertase inputs. However, we realize that the user is most likely not interested in the entire combination/permutation output space. Due to this, we propose a top-down design approach to a tailored reconfigurable circuit. | ||
| + | <p> | ||
| + | If the user is not interested in the entire permutation/combination space, many of the att sites can be removed from the fully reconfigurable design. Thus, Trumpet follows the following process: First, the user specifies the permutations/combinations of interest. Then, based on which algorithm is chosen (Pancake or LinkSort), a fully reconfigurable circuit is produced with att sites that allow for all permutations and combinations. Lastly, all of the att sites which are unnecessary for achieving the desired permutations and combinations are removed. The output of this is a tailored design with only the att sites necessary for the user’s desired permutations/combinations. We call this entire process the top-down design approach. | ||
| + | <p> | ||
| + | We hope that by attempting to solve this problem, we might inspire others out there to design their own att site placing algorithms which are more efficient than ours. Or, perhaps, people interested in this problem can find new and exciting ways to reconfigure circuit plasmids. We believe that if this problem could be solved very efficiently, it might pave the way to more interesting genetic circuits. | ||
| + | </div> | ||
| + | <div id="example"> | ||
| + | <h1>Design Example</h1> | ||
| + | <p> | ||
| + | <img style="float:right; width:320px" src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/Trumpet/twoPartDesign.png"/> | ||
| + | To the right, we see a solution for a two part design which helps illustrate the use of invertases for re configuring a plasmid. This circuit plasmid allows for all permutations/combinations of the two genes using only three sets of invertase sites, illustrated by the sets of red, blue, and orange triangles. By flanking each gene with its own set of att sites, each gene can be turned "on" and "off" by inputting the corresponding invertase. To change the order of the two genes, the invertase corresponding to the outermost att sites (the red sites) must be inputted into the system. The cartoon to the right displays a sequence of invertase inputs, and the effect each input has on the circuit plasmid. The order of these inputs does not particularly matter in this example; however, in more complicated designs the sequence is very important. Also, it should be noted that only four out of the eight possible configurations of this design are displayed in the cartoon. | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div id ="download"> | ||
| + | <h1>Tutorial and Download</h1> | ||
| + | <p> | ||
| + | <a href="https://github.com/igemsoftware/BU_Wellesley_Software_2011/tree/master/Trumpet"><img id="download_button" src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/temp_download_button.jpg" width="100px"/></a> | ||
| + | <p> | ||
| + | <center> | ||
| + | <iframe width="425" height="349" src="http://www.youtube.com/embed/B8_3RUEwY_8?hl=en&fs=1" frameborder="0" allowfullscreen></iframe></center> | ||
<br><br> | <br><br> | ||
| + | </div> | ||
| - | + | <div id="tutorial"> | |
| + | <h1>Tabletop Implementation</h1> | ||
| + | <p>After seeing the success of the desktop implementation of Trumpet, we realized it could be useful to have a tabletop version of Trumpet to allow for collaboration between users. The process of choosing specific combinations of genes, promoters, ribosome binding sites, and other genomic sequences inherently relies on collaboration, since the outcome of such a process will influence the work that each member of the lab needs to complete. Different members of the lab will have different research goals for their work, and the best way to figure out how each person will accomplish their goals in the context of the overall research question requires input from the entire team. A tabletop Trumpet application will allow the entire lab to get involved in determining research goals. | ||
| - | + | <p>To this end, we created and iterated upon a design for a tabletop version of Trumpet. After some initial discussions in the computational team, we showed our goals to the wetlab researchers and got their feedback on which features they would find useful in a tabletop version. While we were not able to create the tabletop version of Trumpet this year due to time constraints, we anticipate adding it to the suite of Surface tools in the future. | |
| + | <br> | ||
| + | <center><iframe width="425" height="349" src="http://www.youtube.com/embed/AGQ9VbdOIeM" frameborder="0" allowfullscreen></iframe></center> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| - | |||
| - | We | + | <div id="results"> |
| + | <h1>Results</h1> | ||
| + | <p> | ||
| + | After designing the primers for the TB transcription factors and sequencing these genes, we can build our constructs with the genes of interest and invertase sites using Trumpet's invertase tree. The tree helps us visualize when we need to apply each invertase to the genetic device. We will be using PCR-based methodologies to add the invertase sites and the transcription factors to a biobrick backbone. This way we can restriction digest these parts and ligate them to the basic devices we constructed. | ||
| + | <p> | ||
| + | In order for the correct permutation to take place, we need to treat the complete construct with invertases. The invertase tree will be useful in telling us the order in which we need to treat the device with the appropriate invertases. The first invertase will be on a seperate plasmid then during transformation of the larger construct we will heat shock the construct to prompt the invertase's actions on the construct. This step is will activate the inversion and flipping action of the desired permutation. With these steps, we will be able to manipulate the actions of the promoter on a gene or even investigate the relationships between a variety of TB transcription factors in a network. | ||
| + | <p> | ||
| + | <center><img style="float:center; width:600px; height:370px" id="invertase_map" src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/Wet_Lab/InvertaseKeyTree.png"></center> | ||
| + | <p> | ||
| - | |||
| - | + | <div id="futurework"> | |
| + | <h1>Future Work</h1> | ||
| + | <p> | ||
| + | <ul> | ||
| + | <li>A surface application for Trumpet is still being developed. On the surface, users would be able to directly manipulate and arrange parts and participate in collaborative design and decision making. Design prototypes include integration with the Clotho database and ability to drag and drop components from a toolbox of parts. The patterns generated by Trumpet could then be translated for use in the lab with <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow </a>and further integration with other applications. | ||
| + | <li>The two algorithms we developed have different required invertase complexities which scale with the number of genes,n. The "better" of these two algorithms has an O(n<sup>2</sup>) complexity. We believe that lower complexity algorithms may exist. Future versions of Trumpet will implement more efficient att site placing algorithms, possibly using a bottom-up approach rather than a top-down approach. | ||
| + | <li>Each of Trumpet's algorithms has an associated key generator. For any permutation, the key generator returns the proper key according to the logic behind the algorithm. While the keys returned are certainly correct, they are not guaranteed to be the shortest possible keys. As the complexity scales with n<sup>2</sup>, we can imagine that some invertase sites which were included in the design for permutation A, for example, could also be used for permutation B. Unfortunately, the key generator has no way of knowing that these sites would also be useful for permutation B. Intelligent algorithms should be developed to find these minimum length keys. | ||
| + | <li>Future versions of Trumpet should also allow the user to specify the maximum length of the longest key, as this may directly correspond to the number of vectors added to the cell. By specifying this length, future Trumpet could return a set of designs, each individually adhering to this maximum key length. As a set, these designs still achieve all desired permutations/combinations. Our current version of Trumpet allows the user to split a design into small sub-designs in an attempt to address this feature. | ||
| + | </ul> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div><!--end bu-wellesley_wiki_content div--> | </div><!--end bu-wellesley_wiki_content div--> | ||
Latest revision as of 01:06, 29 September 2011
Trumpet
Tool Overview
We are developing a synthetic testbed design for a DNA molecule, which we call Trumpet. A trumpet testbed comprises a library of genes and promoters, which can be configured into any desired permutation or combination by treating the DNA with a sequence of enzymes called recombinases. Our design algorithm takes a set of DNA parts, and produces a testbed design and enzyme key dictionary, such that the testbed can be configured into any sequence of promoters and genes by treating it with the corresponding enzyme key. This allows us to rewire the genes and switches and study all their combinations.
 The goal of Trumpet is to aid the user in designing fully reconfigurable genetic circuit plasmids. Imagine that as a researcher, you have a particular set of genes, and you are interested in how the expressions of these genes interact with one another. Conventionally, in order to do this, you must tediously construct many different plasmids, each with the correct combinations and permutations of these genes, and study the expressions of these plasmids one at a time.
The goal of Trumpet is to aid the user in designing fully reconfigurable genetic circuit plasmids. Imagine that as a researcher, you have a particular set of genes, and you are interested in how the expressions of these genes interact with one another. Conventionally, in order to do this, you must tediously construct many different plasmids, each with the correct combinations and permutations of these genes, and study the expressions of these plasmids one at a time.
However, what if you could make a single plasmid containing the genes of interest, and use enzymatic reactions to reconfigure the genes into the different permutations and combinations? Not only would this save a great deal of time in the building phase, but it would open the door to a world of completely reconfigurable genetic circuits based on user input via recombinase embedded suicide vectors. Trumpet is our first attempt at this dream. We have developed a software tool that takes in a set of genes along with the user’s desired set of permutations/combinations, and returns to the user an intelligently designed plasmid which can be reconfigured into the permutations/combinations specified by the user. Along with the plasmid design, Trumpet outputs a set of “keys” for each permutation/combination. These keys indicate the ordered set of recombinase inputs which must be introduced by the user in order to reconfigure the plasmid into the proper state. By developing Trumpet as a Clotho application, the user can quickly save this design to their database of parts. Then, they can use Puppetshow, another one of our applications, to quickly and efficiently build the plasmid using a liquid handling robot.
Method
For this project, we focused on the use of invertases as our recombinase inputs. By orienting two identical copies of an att site in opposite directions, the corresponding invertase which recognizes these att sites will effectively flip the entire duplex region located between the sites. Using this property, we developed two separate algorithms which place at sites intelligently between and around the genes of interest, such that all of the desired combinations and permutations can be achieved through these enzymatic inversions.
The output of our Pancake and LinkSort translations is a large composite part-containing set of genes with att sites properly inserted such that all permutations and combinations of the genes can be achieved given a particular set of invertase inputs. However, we realize that the user is most likely not interested in the entire combination/permutation output space. Due to this, we propose a top-down design approach to a tailored reconfigurable circuit.
If the user is not interested in the entire permutation/combination space, many of the att sites can be removed from the fully reconfigurable design. Thus, Trumpet follows the following process: First, the user specifies the permutations/combinations of interest. Then, based on which algorithm is chosen (Pancake or LinkSort), a fully reconfigurable circuit is produced with att sites that allow for all permutations and combinations. Lastly, all of the att sites which are unnecessary for achieving the desired permutations and combinations are removed. The output of this is a tailored design with only the att sites necessary for the user’s desired permutations/combinations. We call this entire process the top-down design approach.
We hope that by attempting to solve this problem, we might inspire others out there to design their own att site placing algorithms which are more efficient than ours. Or, perhaps, people interested in this problem can find new and exciting ways to reconfigure circuit plasmids. We believe that if this problem could be solved very efficiently, it might pave the way to more interesting genetic circuits.
Design Example
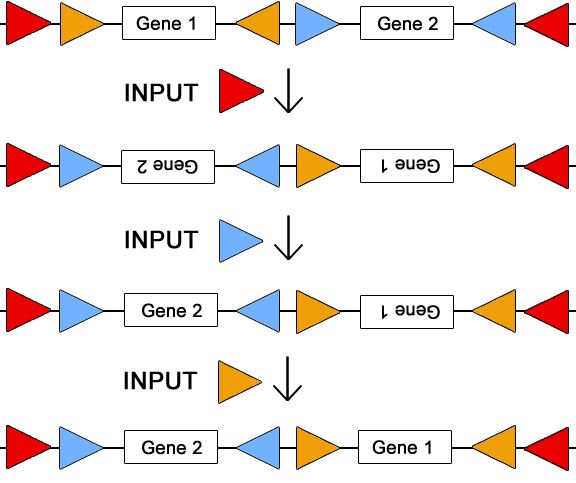 To the right, we see a solution for a two part design which helps illustrate the use of invertases for re configuring a plasmid. This circuit plasmid allows for all permutations/combinations of the two genes using only three sets of invertase sites, illustrated by the sets of red, blue, and orange triangles. By flanking each gene with its own set of att sites, each gene can be turned "on" and "off" by inputting the corresponding invertase. To change the order of the two genes, the invertase corresponding to the outermost att sites (the red sites) must be inputted into the system. The cartoon to the right displays a sequence of invertase inputs, and the effect each input has on the circuit plasmid. The order of these inputs does not particularly matter in this example; however, in more complicated designs the sequence is very important. Also, it should be noted that only four out of the eight possible configurations of this design are displayed in the cartoon.
To the right, we see a solution for a two part design which helps illustrate the use of invertases for re configuring a plasmid. This circuit plasmid allows for all permutations/combinations of the two genes using only three sets of invertase sites, illustrated by the sets of red, blue, and orange triangles. By flanking each gene with its own set of att sites, each gene can be turned "on" and "off" by inputting the corresponding invertase. To change the order of the two genes, the invertase corresponding to the outermost att sites (the red sites) must be inputted into the system. The cartoon to the right displays a sequence of invertase inputs, and the effect each input has on the circuit plasmid. The order of these inputs does not particularly matter in this example; however, in more complicated designs the sequence is very important. Also, it should be noted that only four out of the eight possible configurations of this design are displayed in the cartoon.
Tabletop Implementation
After seeing the success of the desktop implementation of Trumpet, we realized it could be useful to have a tabletop version of Trumpet to allow for collaboration between users. The process of choosing specific combinations of genes, promoters, ribosome binding sites, and other genomic sequences inherently relies on collaboration, since the outcome of such a process will influence the work that each member of the lab needs to complete. Different members of the lab will have different research goals for their work, and the best way to figure out how each person will accomplish their goals in the context of the overall research question requires input from the entire team. A tabletop Trumpet application will allow the entire lab to get involved in determining research goals.
To this end, we created and iterated upon a design for a tabletop version of Trumpet. After some initial discussions in the computational team, we showed our goals to the wetlab researchers and got their feedback on which features they would find useful in a tabletop version. While we were not able to create the tabletop version of Trumpet this year due to time constraints, we anticipate adding it to the suite of Surface tools in the future.
Results
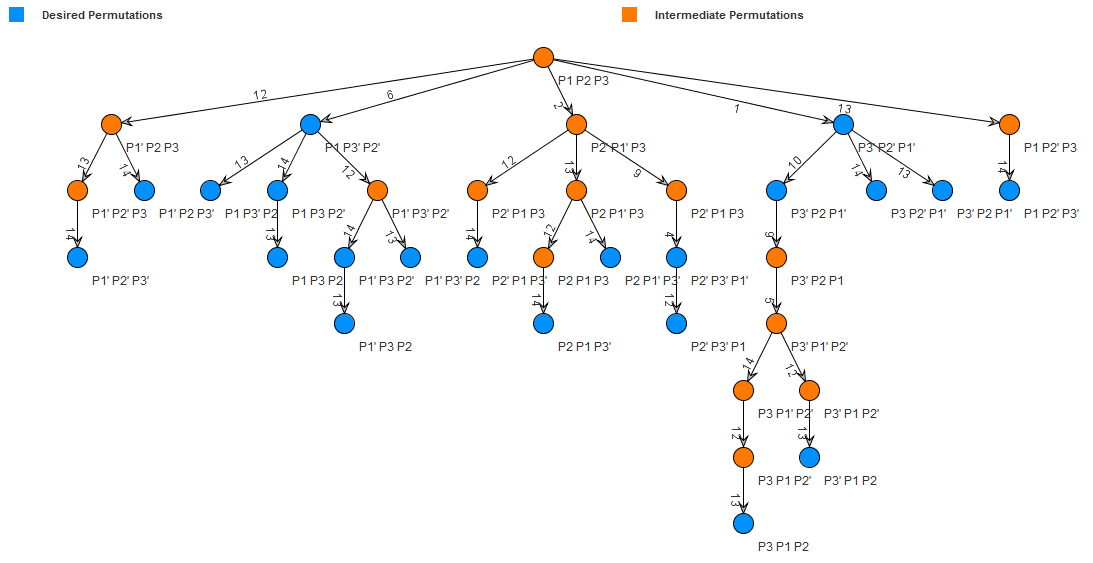
After designing the primers for the TB transcription factors and sequencing these genes, we can build our constructs with the genes of interest and invertase sites using Trumpet's invertase tree. The tree helps us visualize when we need to apply each invertase to the genetic device. We will be using PCR-based methodologies to add the invertase sites and the transcription factors to a biobrick backbone. This way we can restriction digest these parts and ligate them to the basic devices we constructed.
In order for the correct permutation to take place, we need to treat the complete construct with invertases. The invertase tree will be useful in telling us the order in which we need to treat the device with the appropriate invertases. The first invertase will be on a seperate plasmid then during transformation of the larger construct we will heat shock the construct to prompt the invertase's actions on the construct. This step is will activate the inversion and flipping action of the desired permutation. With these steps, we will be able to manipulate the actions of the promoter on a gene or even investigate the relationships between a variety of TB transcription factors in a network.
Future Work
- A surface application for Trumpet is still being developed. On the surface, users would be able to directly manipulate and arrange parts and participate in collaborative design and decision making. Design prototypes include integration with the Clotho database and ability to drag and drop components from a toolbox of parts. The patterns generated by Trumpet could then be translated for use in the lab with Puppetshow and further integration with other applications.
- The two algorithms we developed have different required invertase complexities which scale with the number of genes,n. The "better" of these two algorithms has an O(n2) complexity. We believe that lower complexity algorithms may exist. Future versions of Trumpet will implement more efficient att site placing algorithms, possibly using a bottom-up approach rather than a top-down approach.
- Each of Trumpet's algorithms has an associated key generator. For any permutation, the key generator returns the proper key according to the logic behind the algorithm. While the keys returned are certainly correct, they are not guaranteed to be the shortest possible keys. As the complexity scales with n2, we can imagine that some invertase sites which were included in the design for permutation A, for example, could also be used for permutation B. Unfortunately, the key generator has no way of knowing that these sites would also be useful for permutation B. Intelligent algorithms should be developed to find these minimum length keys.
- Future versions of Trumpet should also allow the user to specify the maximum length of the longest key, as this may directly correspond to the number of vectors added to the cell. By specifying this length, future Trumpet could return a set of designs, each individually adhering to this maximum key length. As a set, these designs still achieve all desired permutations/combinations. Our current version of Trumpet allows the user to split a design into small sub-designs in an attempt to address this feature.
 "
"

