Team:BU Wellesley Software/Trumpet
From 2011.igem.org
| Line 97: | Line 97: | ||
To develop these algorithms, we recognized this problem as a special case of sorting which only allows for inversions in order to sort a set of elements. Thus, our first step towards solving this sorting problem was to develop sorting algorithms that follow these inversion rules. The result of this process was two algorithms which we have called “Pancake” and “LinkSort” (due to their sorting nature). While one can imagine any number of ways to sort elements through inversions, the next step, and certainly the more difficult and important step, was to translate these algorithms into a set of rules which intelligently places att sites such that, by following the corresponding algorithm, there is always a set of att sites available for the next inversion. | To develop these algorithms, we recognized this problem as a special case of sorting which only allows for inversions in order to sort a set of elements. Thus, our first step towards solving this sorting problem was to develop sorting algorithms that follow these inversion rules. The result of this process was two algorithms which we have called “Pancake” and “LinkSort” (due to their sorting nature). While one can imagine any number of ways to sort elements through inversions, the next step, and certainly the more difficult and important step, was to translate these algorithms into a set of rules which intelligently places att sites such that, by following the corresponding algorithm, there is always a set of att sites available for the next inversion. | ||
<p> | <p> | ||
| - | The output of our Pancake and LinkSort translations is a large composite part-containing set of genes with att sites properly inserted such that all permutations and combinations of the genes can be achieved given a particular set of invertase inputs. However, we realize that the user is most likely not interested in the entire combination/permutation output space. Due to this, we propose a top-down design approach to a tailored reconfigurable circuit. | + | The output of our Pancake and LinkSort translations is a large composite part-containing set of genes with att sites properly inserted such that all permutations and combinations of the genes can be achieved given a particular set of invertase inputs. However, we realize that the user is most likely not interested in the entire combination/permutation output space. Due to this, we propose a top-down design approach to a tailored reconfigurable circuit. |
<p> | <p> | ||
If the user is not interested in the entire permutation/combination space, many of the att sites can be removed from the fully reconfigurable design. Thus, Trumpet follows the following process: First, the user specifies the permutations/combinations of interest. Then, based on which algorithm is chosen (Pancake or LinkSort), a fully reconfigurable circuit is produced with att sites that allow for all permutations and combinations. Lastly, all of the att sites which are unnecessary for achieving the desired permutations and combinations are removed. The output of this is a tailored design with only the att sites necessary for the user’s desired permutations/combinations. We call this entire process the top-down design approach. | If the user is not interested in the entire permutation/combination space, many of the att sites can be removed from the fully reconfigurable design. Thus, Trumpet follows the following process: First, the user specifies the permutations/combinations of interest. Then, based on which algorithm is chosen (Pancake or LinkSort), a fully reconfigurable circuit is produced with att sites that allow for all permutations and combinations. Lastly, all of the att sites which are unnecessary for achieving the desired permutations and combinations are removed. The output of this is a tailored design with only the att sites necessary for the user’s desired permutations/combinations. We call this entire process the top-down design approach. | ||
| Line 104: | Line 104: | ||
<p> | <p> | ||
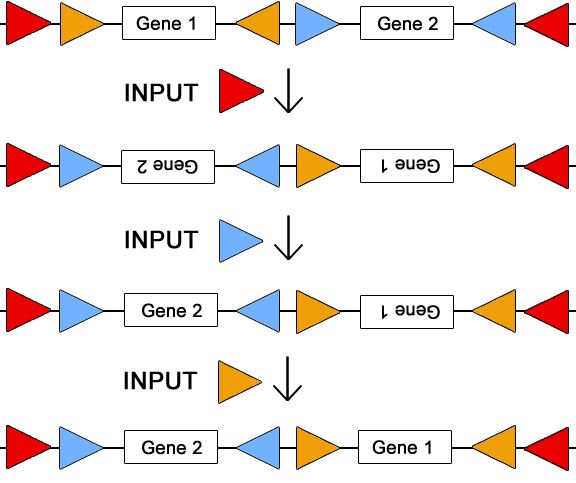
Here we explain a simple, yet nontrivial, solution for a two part design which helps illustrate the use of invertases for re configuring a plasmid. This circuit plasmid allows for all permutations/combinations of the two genes using only three sets of invertase sites, illustrated by the sets of red, blue, and orange triangles. By flanking each gene with a its own set of att sites, each gene can be turned "on" and "off" by inputting the corresponding invertase. To change the order of the two genes, the invertase corresponding to the outermost att sites (the red sites) must be inputted into the system. Below is a cartoon displaying the sequence of inputs that leads to three of the five total permutations/combinations. | Here we explain a simple, yet nontrivial, solution for a two part design which helps illustrate the use of invertases for re configuring a plasmid. This circuit plasmid allows for all permutations/combinations of the two genes using only three sets of invertase sites, illustrated by the sets of red, blue, and orange triangles. By flanking each gene with a its own set of att sites, each gene can be turned "on" and "off" by inputting the corresponding invertase. To change the order of the two genes, the invertase corresponding to the outermost att sites (the red sites) must be inputted into the system. Below is a cartoon displaying the sequence of inputs that leads to three of the five total permutations/combinations. | ||
| - | <img style="width: | + | <p> |
| + | <img style="width:350px; padding:10px;" align="middle" src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/Trumpet/twoPartDesign.png"/> | ||
<p> | <p> | ||
<b>Download: </b><br> | <b>Download: </b><br> | ||
| - | <a href="#"><img id="download_button" src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/temp_download_button.jpg"/></a> | + | <a href="#"><img id="download_button" src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/temp_download_button.jpg" width="100px"/></a> |
<p> | <p> | ||
<b>Demo Video: </b> | <b>Demo Video: </b> | ||
| Line 125: | Line 126: | ||
<h1>Future Work</h1> | <h1>Future Work</h1> | ||
<p> | <p> | ||
| - | A surface application for Trumpet is still being developed. On the surface, users would be able to directly manipulate and arrange parts and participate in collaborative design and decision making. Design prototypes include integration with the Clotho database and ability to drag and drop components from a toolbox of parts. The patterns generated by Trumpet could then be translated for use in the lab with <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow </a>and further integration with other applications. | + | <ul> |
| + | <li>A surface application for Trumpet is still being developed. On the surface, users would be able to directly manipulate and arrange parts and participate in collaborative design and decision making. Design prototypes include integration with the Clotho database and ability to drag and drop components from a toolbox of parts. The patterns generated by Trumpet could then be translated for use in the lab with <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow </a>and further integration with other applications. | ||
| + | </ul> | ||
| + | |||
</div> | </div> | ||
Revision as of 21:47, 12 September 2011
Trumpet
Tool Overview
We are developing a synthetic testbed design for a DNA molecule, which we call Trumpet. A trumpet testbed comprises a library of genes and promoters, which can be configured into any desired permutation or combination by treating the DNA with a sequence of enzymes called recombinases. Our design algorithm takes a set of DNA parts, and produces a testbed design and enzyme key dictionary, such that the testbed can be configured into any sequence of promoters and genes by treating it with the corresponding enzyme key. This allows us to rewire the genes and switches and study all their combinations.
The goal of Trumpet is to aid the user in designing fully reconfigurable genetic circuit plasmids. Imagine that as a researcher, you have a particular set of genes, and you are interested in how the expressions of these genes interact with one another. Conventionally, in order to do this, you must tediously construct many different plasmids, each with the correct combinations and permutations of these genes, and study the expressions of these plasmids one at a time.

However, what if you could make a single plasmid containing the genes of interest, and use enzymatic reactions to reconfigure the genes into the different permutations and combinations? Not only would this save a great deal of time in the building phase, but it would open the door to a world of completely reconfigurable genetic circuits based on user input via recombinase embedded suicide vectors. Trumpet is our first attempt at this dream. We have developed a software tool that takes in a set of genes along with the user’s desired set of permutations/combinations, and returns to the user an intelligently designed plasmid which can be reconfigured into the permutations/combinations specified by the user. Along with the plasmid design, Trumpet outputs a set of “keys” for each permutation/combination. These keys indicate the ordered set of recombinase inputs which must be introduced by the user in order to reconfigure the plasmid into the proper state. By developing Trumpet as a Clotho application, the user can quickly save this design to their database of parts. Then, they can use Puppetshow, another one of our applications, to quickly and efficiently build the plasmid using a liquid handling robot.
Design Method: (CL: This needs to show some configurable constructs that were designed using Trumpet.) For this project, we focused on the use of invertases as our recombinase inputs. By orienting two identical copies of an att site in opposite directions, the corresponding invertase which recognizes these att sites will effectively flip the entire duplex region located between the sites. Using this property, we developed two separate algorithms which place at sites intelligently between and around the genes of interest, such that all of the desired combinations and permutations can be achieved through these enzymatic inversions.
To develop these algorithms, we recognized this problem as a special case of sorting which only allows for inversions in order to sort a set of elements. Thus, our first step towards solving this sorting problem was to develop sorting algorithms that follow these inversion rules. The result of this process was two algorithms which we have called “Pancake” and “LinkSort” (due to their sorting nature). While one can imagine any number of ways to sort elements through inversions, the next step, and certainly the more difficult and important step, was to translate these algorithms into a set of rules which intelligently places att sites such that, by following the corresponding algorithm, there is always a set of att sites available for the next inversion.
The output of our Pancake and LinkSort translations is a large composite part-containing set of genes with att sites properly inserted such that all permutations and combinations of the genes can be achieved given a particular set of invertase inputs. However, we realize that the user is most likely not interested in the entire combination/permutation output space. Due to this, we propose a top-down design approach to a tailored reconfigurable circuit.
If the user is not interested in the entire permutation/combination space, many of the att sites can be removed from the fully reconfigurable design. Thus, Trumpet follows the following process: First, the user specifies the permutations/combinations of interest. Then, based on which algorithm is chosen (Pancake or LinkSort), a fully reconfigurable circuit is produced with att sites that allow for all permutations and combinations. Lastly, all of the att sites which are unnecessary for achieving the desired permutations and combinations are removed. The output of this is a tailored design with only the att sites necessary for the user’s desired permutations/combinations. We call this entire process the top-down design approach.
We hope that by attempting to solve this problem, we might inspire others out there to design their own att site placing algorithms which are more efficient than ours. Or, perhaps, people interested in this problem can find new and exciting ways to reconfigure circuit plasmids. We believe that if this problem could be solved very efficiently, it might pave the way to more interesting genetic circuits.
Here we explain a simple, yet nontrivial, solution for a two part design which helps illustrate the use of invertases for re configuring a plasmid. This circuit plasmid allows for all permutations/combinations of the two genes using only three sets of invertase sites, illustrated by the sets of red, blue, and orange triangles. By flanking each gene with a its own set of att sites, each gene can be turned "on" and "off" by inputting the corresponding invertase. To change the order of the two genes, the invertase corresponding to the outermost att sites (the red sites) must be inputted into the system. Below is a cartoon displaying the sequence of inputs that leads to three of the five total permutations/combinations.
Demo Video:
Results
Wetlab content goes here.
Future Work
- A surface application for Trumpet is still being developed. On the surface, users would be able to directly manipulate and arrange parts and participate in collaborative design and decision making. Design prototypes include integration with the Clotho database and ability to drag and drop components from a toolbox of parts. The patterns generated by Trumpet could then be translated for use in the lab with Puppetshow and further integration with other applications.
 "
"

