Team:BU Wellesley Software/Notebook/CaseyNotebook
From 2011.igem.org
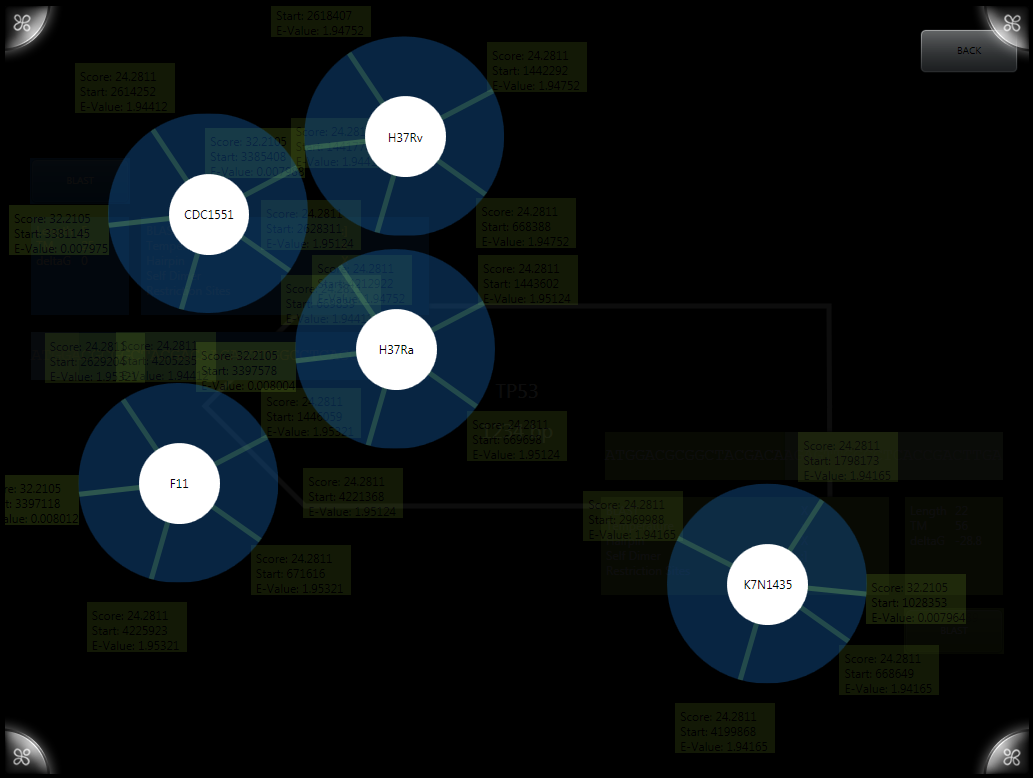
This summer the Wellesley HCI Lab worked on re-implementing an old system, G-nome Surfer 2.0, new and improved and optimized for prokaryotic genomes. To make an application practical for researching prokaryotic genomes we had to create a number of new interaction styles for dealing with navigation of a circular chromosome. Working on the system I worked on the new chromosome wheel, the publications system, the new primer designer and the associated BLAST.
Implementation G-Nome Surfer Pro is written in C# using the Microsoft SDK for use on the Microsoft Surface. After much trial and error we re-implemented many parts of the system using data binding which allows a complete separation of state and logic. (more information about data binding can be found on MSDN [http://msdn.microsoft.com/en-us/library/ms752347.aspx here])
Navigation The central chromosome wheel went through several iterations before arriving at the final simple rotation model we currently use. An early implementation allowed for zooming through use of a pair of arms (like in the early paper prototype.) Like many of the early user controls the arms involved a fair amount of hacking through the presentation logic and item templates through the .xaml files.
Publications
Primer Design
BLASTing
 "
"