Team:Arizona State/Project
From 2011.igem.org
Rubenacuna (Talk | contribs) (added diagram 2) |
Rubenacuna (Talk | contribs) (added in text link to diagram 2) |
||
| Line 11: | Line 11: | ||
'''The
CRISPR
Mechanism
''' | '''The
CRISPR
Mechanism
''' | ||
---- | ---- | ||
| - |
The
CRISPR‐Cas
pathway
can
be
compared
to
a
prokaryotic
immunity
or
RNA
interference
that
can
be
directed
to
silence
a
gene
of
interest.
This
mechanism
of
bacterial
survival
affords
us
an
interesting
method
to
tackle
the
aforementioned
problem.
Clustered
Regularly
Interspaced
Short
Palindromic
Repeats
(CRISPR)
gene
loci
have
been
demonstrated
to
equip
both
prokaryotes
and
archaea
with
a
defense
mechanism
against
exogenous
DNA
and
RNA
sequences [[#ref1| [1]]], [[#ref2| [2]]].
CRISPR
genes
appear
in
an
array
that
contains
contiguous
spacers,
repeats,
and
an
operon
of
structural
genes.
The
transcripts
from
the
spacer/repeat
region
undergo
hair
pinning
due
to
the
palindromic
sequence
structure.
The
peptide
products
of
the
CRISPR‐associated
structural
genes
(CAS)
work
cooperatively
with
crRNA
to
silence
a
complimentary
target
[[#diagram1| (Diagram 1)]] [[#ref3| [3]]]. The
function
is
a
prokaryotic
analog
to
both
RNA
interference
and
immunity.
CRISPR
quickly
presents
it self
as
a
potentially
useful
tool
in
prokaryotic
gene
manipulation.
Our
goal
as
ASU’s
first
iGEM
team
is
to
develop
a
CRISPR
plasmid
that
contains
elements
to
target
and
silence
the
NDM‐1
gene
sequence
( | + |
The
CRISPR‐Cas
pathway
can
be
compared
to
a
prokaryotic
immunity
or
RNA
interference
that
can
be
directed
to
silence
a
gene
of
interest.
This
mechanism
of
bacterial
survival
affords
us
an
interesting
method
to
tackle
the
aforementioned
problem.
Clustered
Regularly
Interspaced
Short
Palindromic
Repeats
(CRISPR)
gene
loci
have
been
demonstrated
to
equip
both
prokaryotes
and
archaea
with
a
defense
mechanism
against
exogenous
DNA
and
RNA
sequences [[#ref1| [1]]], [[#ref2| [2]]].
CRISPR
genes
appear
in
an
array
that
contains
contiguous
spacers,
repeats,
and
an
operon
of
structural
genes.
The
transcripts
from
the
spacer/repeat
region
undergo
hair
pinning
due
to
the
palindromic
sequence
structure.
The
peptide
products
of
the
CRISPR‐associated
structural
genes
(CAS)
work
cooperatively
with
crRNA
to
silence
a
complimentary
target
[[#diagram1| (Diagram 1)]] [[#ref3| [3]]]. The
function
is
a
prokaryotic
analog
to
both
RNA
interference
and
immunity.
CRISPR
quickly
presents
it self
as
a
potentially
useful
tool
in
prokaryotic
gene
manipulation.
Our
goal
as
ASU’s
first
iGEM
team
is
to
develop
a
CRISPR
plasmid
that
contains
elements
to
target
and
silence
the
NDM‐1
gene
sequence
[[#diagram2| (Diagram 2)]].
While
targeting
NDM‐1,
we
recognize
that
CRISPR can
potentially
target
any
gene
of
interest,
thus
we
will
develop
a
robust
platform
for
gene
silencing.
The
final
product
of
this
project
will
be
a
fully
functioning
CRISPR
array
that
will
be
submitted
to
the
Standard
Registry
of
Biological
Parts,
an
open‐source
collection
of
DNA
building
blocks,
as
a
BioBrick,
a
modular
component
for
genetic
engineering
[[#diagram3| (Diagram 3)]].
|
<div id="diagram1"></div> | <div id="diagram1"></div> | ||
Revision as of 18:56, 11 June 2011
Project
- Our project will have several stages, all pursuant to the general investigation and modularization of the CRISPR pathway:
- Proof of concept targeting reporters such as GFP, eventually creating a CRISPR biobrick
- Investigate CRISPR system dynamics based on factors such as degradation of self-targeting sequences and maintenance of the array.
- Target genes such as NDM-1 or other clinically relevant pathways.
NDM-1 in Perspective
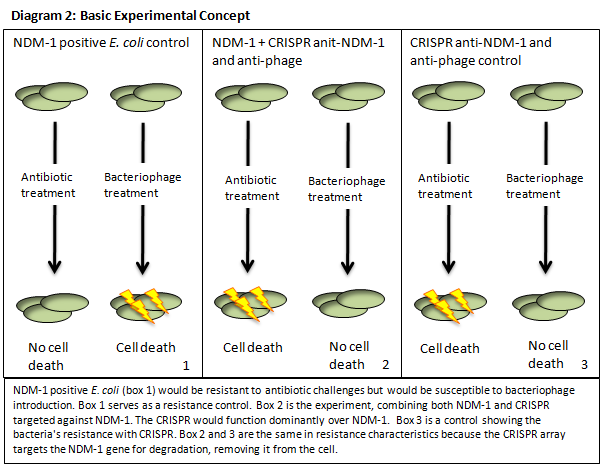
Global antibiotic resistance is a concern of the utmost importance to the World Health Organization and health care everywhere. Bacteria that have acquired antibiotic resistance jeopardize world health care as a whole, because they increase mortality rate of normally curable infections, and there is no coherent approach to containing and countering resistant strains. New Delhi Metallo‐Beta‐Lactamse (NDM‐1) containing bacteria are particularly ominous because the NDM‐1 enzyme hydrolyzes a broad range of potent beta‐lactam antibiotics (e.g. carbapenems). This enzyme is effective in rendering normal lines of treatment for bacterial infection useless. NDM‐1 positive strains originated in India and Pakistan and have recently spread to the UK, Europe, and Canada. There has also been a drastic increase in the number of reported NDM‐1 positive cases in the United States, according to the Centers of Disease Control and Prevention. Viable antibiotics as a resource are becoming more and more deficient. Alternative solutions to resistance must be promptly sought and intelligently employed to counter the threat of antibiotic resistant bacteria.
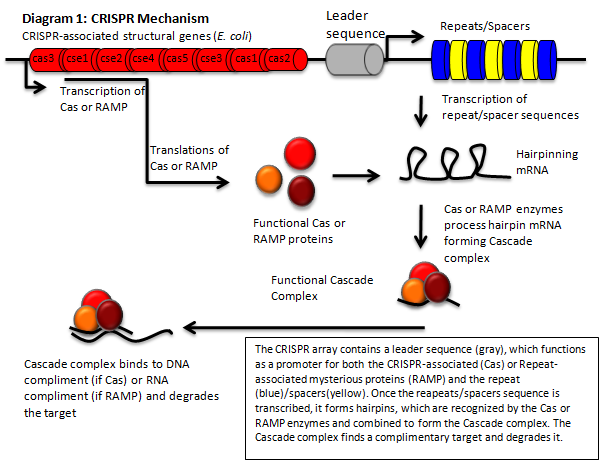
The CRISPR Mechanism
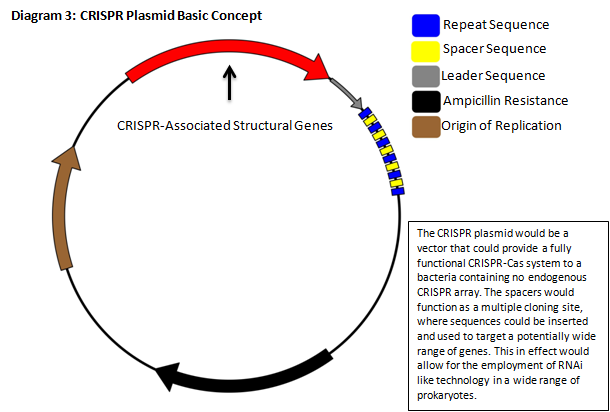
The CRISPR‐Cas pathway can be compared to a prokaryotic immunity or RNA interference that can be directed to silence a gene of interest. This mechanism of bacterial survival affords us an interesting method to tackle the aforementioned problem. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) gene loci have been demonstrated to equip both prokaryotes and archaea with a defense mechanism against exogenous DNA and RNA sequences [1], [2]. CRISPR genes appear in an array that contains contiguous spacers, repeats, and an operon of structural genes. The transcripts from the spacer/repeat region undergo hair pinning due to the palindromic sequence structure. The peptide products of the CRISPR‐associated structural genes (CAS) work cooperatively with crRNA to silence a complimentary target (Diagram 1) [3]. The function is a prokaryotic analog to both RNA interference and immunity. CRISPR quickly presents it self as a potentially useful tool in prokaryotic gene manipulation. Our goal as ASU’s first iGEM team is to develop a CRISPR plasmid that contains elements to target and silence the NDM‐1 gene sequence (Diagram 2). While targeting NDM‐1, we recognize that CRISPR can potentially target any gene of interest, thus we will develop a robust platform for gene silencing. The final product of this project will be a fully functioning CRISPR array that will be submitted to the Standard Registry of Biological Parts, an open‐source collection of DNA building blocks, as a BioBrick, a modular component for genetic engineering (Diagram 3).
References
 "
"