Team:XMU-China/Project/Protocols
From 2011.igem.org
(→Cell growth) |
(→Site Directed Mutagenesis) |
||
| Line 121: | Line 121: | ||
===''' Site Directed Mutagenesis''' === | ===''' Site Directed Mutagenesis''' === | ||
| - | [[Image: | + | [[Image:XMU_China_144.jpg|left]] |
| - | [[Image: | + | [[Image:XMU_China_145.jpg|left]] |
<html> | <html> | ||
<img src="http://partsregistry.org/wiki/images/4/41/XMU_China_block.jpg"> | <img src="http://partsregistry.org/wiki/images/4/41/XMU_China_block.jpg"> | ||
Revision as of 23:49, 5 October 2011
Contents |
Protocols
1.Isolation of Plasmid
Using the Procedure for GenEluteTM Plasmid Miniprep Kit
•Collect 1-5 mL bacterium fluid in 1.5 mL centrifuge tube,and centrifuge fluid at 12000 r/min
•Resuspend cells. Discard the supernatant and completely resuspend the bacterial pellet with 250 µl of the Resuspension Solution
•Lyse cells. Lyse the resuspended cells by adding 250 µl of the Lysis Solution
•Neutralize. Precipitate the cell debris by adding 350 µl of the Neutralization/Binding Solution, and centrifuge fluid at 12000r/min
•Load cleared lysate. Transfer the supernatant from step 4 to the spin column. Centrifuge at 12000r/min for 1 minute, and then discard the filtrate
•Optional wash. Add 500 µl of the Optional Wash Solution to the column. Centrifuge at 12000 r/min for 1 minute. Discard the filtrate
•Wash column. Add 500 µl of the diluted Wash Solution to the column. Centrifuge at 12000r/min for 1 minute.
•Elute DNA. Transfer the column to a new collection tube. Add 50~100 µl of Eluent Solution to the column. Centrifuge at 12000 r/min for 1 minute. The DNA is now present in the filtrate and is ready for immediate use or storage at -20℃
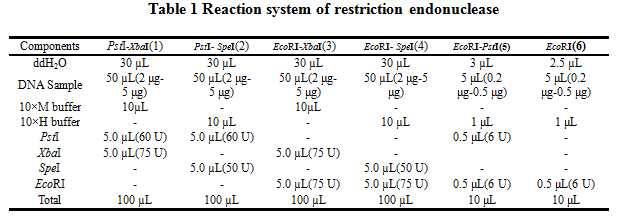
2.Reaction system of restriction endonuclease

System1、2、3 and 4 are used for Standard BioBrick Assembly .System 5 and 6 are used for Restriction analysis. Digestion of sample: at least 500 ng DNA / 10 µL volume. Digest for 4 h at 37 °C, afterwards inactivated by adding 10x loading buffer and standing for 10 min at room temperature.
Standard BioBrick Assembly
•Digestion of insert: 2 μg~5 μg DNA / 100 µL volume, 10x H buffer, EcoRI, SpeI. Digestion and inactivation. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel, try to avoid staining or exposure to ultraviolet light of the insert.
•Digestion of vector: 2 μg~5 μg DNA / 100 µL volume, 10 x M buffer, EcoRI, XbaI. Digestion and inactivation. Clean up the insert via gel electrophoresis. When cutting the insert out of the gel, try to avoid staining or exposure to ultraviolet light of the insert.
Suffix Insertion
•Digestion of insert: 2 μg~5 μg DNA / 100 µL volume, 10x M buffer, XbaI, PstI. Digestion and inactivation. Clean up the insert.
•Digestion of vector : 2 μg~5 μg DNA / 100 µL volume, 10x H buffer, SpeI, PstI. Digestion and inactivation. Clean up the vector.
Ligation
•After digestion and clean-up, the next step is ligation. Overnight ligation at 16°C. Table 2 is the system of ligation.
Transformation
•Preparation of competent E.coli cells
•Add 10 µL plasmid to 100 µl competent cells in centrifuge tube
•Store tube on ice for 20-30 minutes
•Water bath for 90s at 42℃
•Put the tube on ice for 1-2min
•Add 790 µL LB,and cultivation for 1h at 37 ℃,then plate on selective LB-Medium.
Restriction analysis
•Pick one colony with a sterile tip and cultivation in 20ml LB for overnight at 37 ℃
•Isolation of Plasmid
•Digest BioBrick,the system of Restriction analysis refer to table1
•Gel electrophoresis:add 2 µL loading buffer to digestion mixture. An agarose concentration is 1 %.
Determining fluorescence intensity
•Add IPTG when A600 0.6~0.8.
•Cool the culture 10 minutes on ice.
•Centrifuge at 6000rpm. Wash it with pre-cooled PBS buffer.
•Use fluorescence spectrophotometer tomeasure the fluorescenceof GFP:
•Before measuring, dilute the bacteria with PBS buffer so that it can be within the measuring #range. Set excitation wavelength 491 nm, emission wavelength 511 nm.
•Transfer the measured bacteria in a new centrifuge tube and measure the OD of the bacteria.
Cell growth
•100µL suspension was inoculated from a Glycerin tube into 20ml fresh LB and incubated overnight at 37℃ and 250 r.p.m.
•100µL suspension was inoculated again from step1 into 50ml fresh LB and incubated at 37℃ and 250r.p.m
•IPTG was added when A600≈0.6-0.8
•1 ml suspension was taken on every sample taken time. 3 samples were taken in each time.
•Diluted each sample to 10-6(Sometimes 10-5),and then plate on selective LB-Medium.
•After 12h, count the number of CFU on the plate on different time point and then draw the cell growth curve.

Site Directed Mutagenesis

•Digest the template plasmid by adding 1 µL of DpnI and incubate for 1-2 h
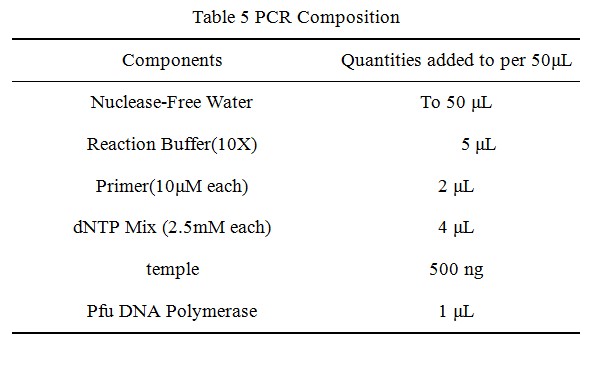
•Transform 10 µL of t PCR product into competent E. coli cells
•Screen the transformants using restiction digest and sequencing
 "
"