Team:Korea U Seoul/Project/Design
From 2011.igem.org
(Difference between revisions)
(→Hydrocarbon detection) |
(→Project Design) |
||
| Line 37: | Line 37: | ||
*Lux genes | *Lux genes | ||
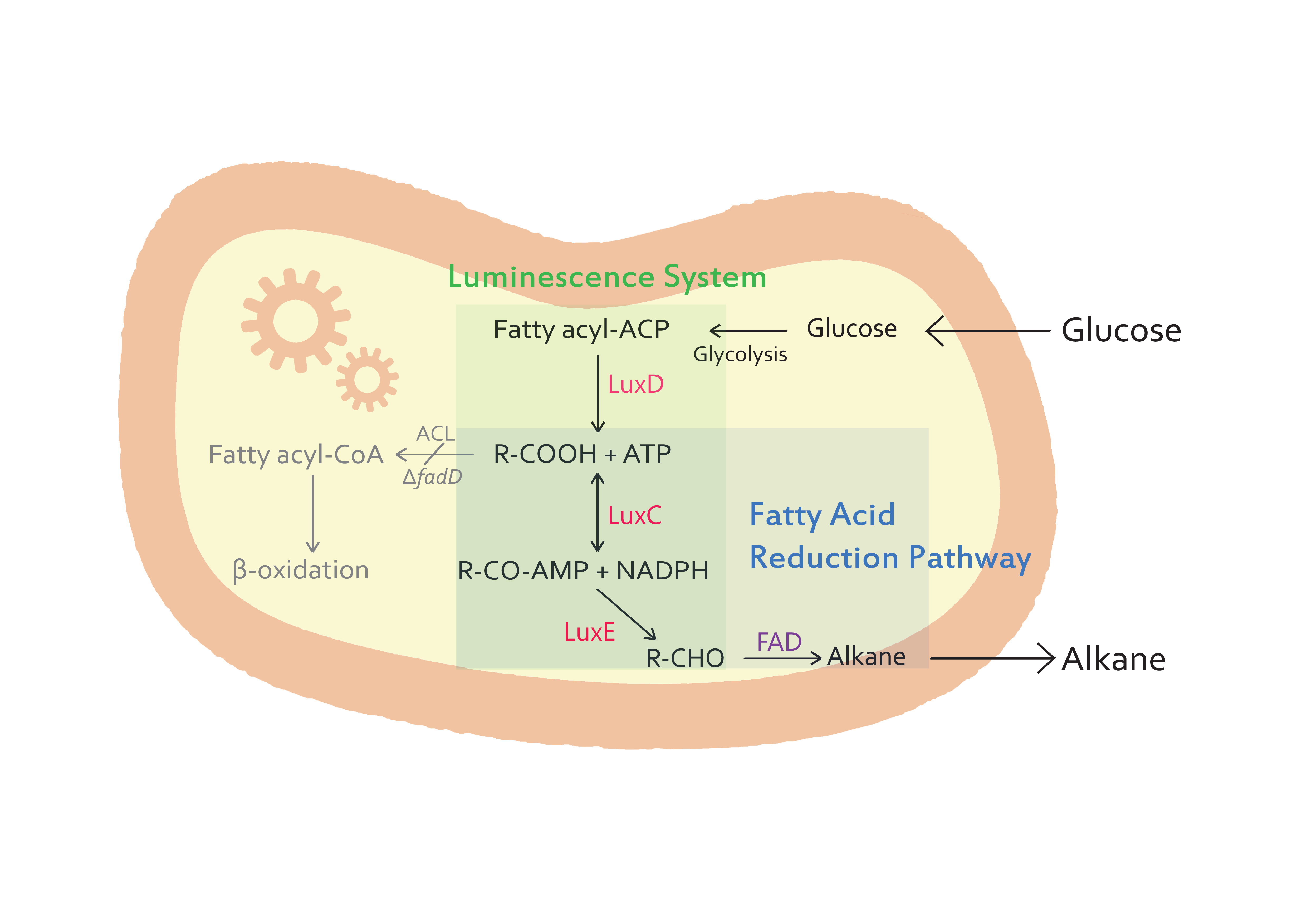
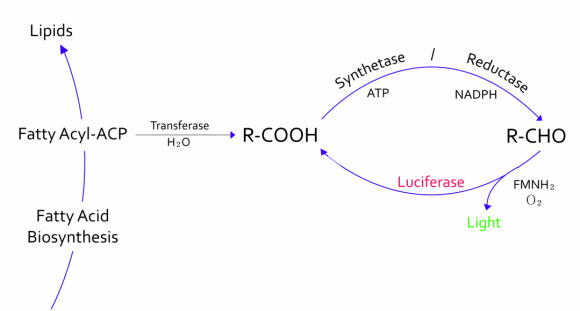
:- The genes from bioluminescence operons have been identified, and we use some structural genes (''luxC'', ''D'', and ''E'' genes). They code for the polypeptides of the fatty acid reductase system responsible for synthesis of the fatty aldehyde substrate. | :- The genes from bioluminescence operons have been identified, and we use some structural genes (''luxC'', ''D'', and ''E'' genes). They code for the polypeptides of the fatty acid reductase system responsible for synthesis of the fatty aldehyde substrate. | ||
| - | + | :- There are 5 lux genes that we used. ''lux A'', ''B'', ''C'', ''D'', and ''E'' were used. ''lux A'' and ''lux B'' codes Luciferase alpha and beta subunit respectively. ''lux C'' codes Reductase, ''lux D'' codes Acyl-transferase, and ''lux E'' codes Synthetase. Luciferase alpha and beta subunit function is catalysis of the bioluminescence reaction(FMNH2 + O2 + aldehyde -> light). Reductase's function is NADPH-dependent reduction of activated fatty acyl groups to aldehyde. Acyl-transferase's function is generation of fatty acids(tetradecanoic acid) for the luminescence system. Lastly, Synthetase's function is ATP-dependent activation of fatty acids.[[File:lux.jpg|thumb|Bioluminescence|500px|center]] | |
| - | + | ||
| - | [[File:lux.jpg|thumb|Bioluminescence|500px|center]] | + | |
[[File:arrows.jpg|thumb|Manipulation of Lux genes|500px|center]] | [[File:arrows.jpg|thumb|Manipulation of Lux genes|500px|center]] | ||
Revision as of 09:55, 5 October 2011
Project Design
- E.coli K27 strains
- - The purpose of our team is to synthesize alkanes from microorganisms. E.coli K27 is a suitable host for the production of alkanes because it is a FadD mutant(△fadD).
- - In our step of alkanes synthesis, the fatty acids are important intermediates. Commonly, E.coli cells contain a single acyl-CoA synthetase, which activates the conversion of free fatty acid to acyl-CoA thioester. However, E.coli K27, a FadD mutant, lacks acyl-CoA synthetase activity, which prevents substrate or product degradation by the host. So, E.coli K27 accumulates fatty acids inside the cell, and finally we can get more alkanes than other E.coli strains.
- Two-carbon compounds and fatty acids as carbon sources
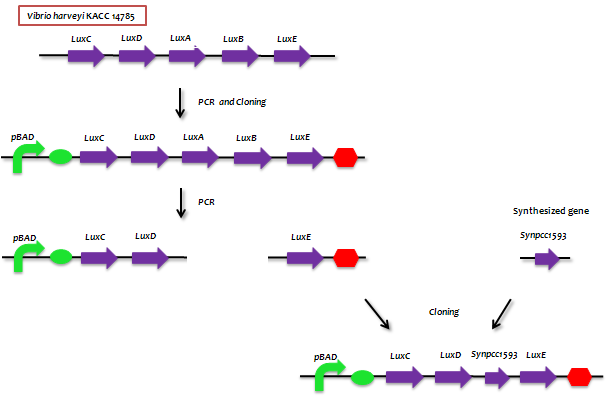
- Lux genes
- - The genes from bioluminescence operons have been identified, and we use some structural genes (luxC, D, and E genes). They code for the polypeptides of the fatty acid reductase system responsible for synthesis of the fatty aldehyde substrate.
- - There are 5 lux genes that we used. lux A, B, C, D, and E were used. lux A and lux B codes Luciferase alpha and beta subunit respectively. lux C codes Reductase, lux D codes Acyl-transferase, and lux E codes Synthetase. Luciferase alpha and beta subunit function is catalysis of the bioluminescence reaction(FMNH2 + O2 + aldehyde -> light). Reductase's function is NADPH-dependent reduction of activated fatty acyl groups to aldehyde. Acyl-transferase's function is generation of fatty acids(tetradecanoic acid) for the luminescence system. Lastly, Synthetase's function is ATP-dependent activation of fatty acids.
Hydrocarbon detection
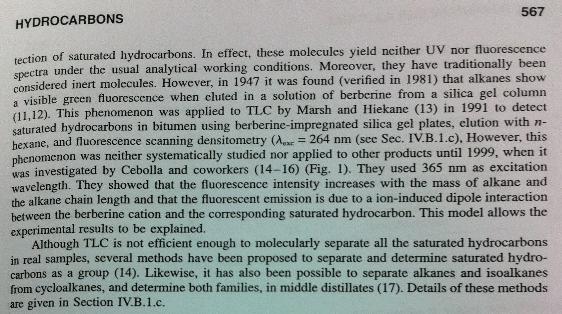
- We are currently having difficulty in detecting the final product, C14.
- - Detection of C14 by TLC is difficult. It is not yet providing good enough results. Apparently, this is due to the fact that saturated hydrocarbons don't react with any other compounds.
- - By incorporating BBa_K32599 in luxCDEG, we may be able to detect hydrocarbon.
Contact : |
synbiogroup@googlegroups.com |
 |
|
|
|
 "
"