Team:WashU/Notebook/July2011
From 2011.igem.org
(→July 20) |
(→July 22) |
||
| Line 300: | Line 300: | ||
*Reverse primer GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT | *Reverse primer GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT | ||
| - | ** | + | **Biobrick: |
*ctgcagcggccgcractagta GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT | *ctgcagcggccgcractagta GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT | ||
| - | |||
**'''CCD1-leu2''' | **'''CCD1-leu2''' | ||
Revision as of 15:34, 25 July 2011
July 1
Master mix composition (6 wells worth)
- 15.0uL 10x buffer
- 7.5 uL dNTP
- 99 uL dH20
- 1.5 uL Accutaq LA DNA polyermerase
= 20.5 ul/well
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 10ng Kan DNA (1mL using our 2x diluted sample)
- 1.5ul of Nat ligation product
2nd attempt at PCR on the Nat ligation product:
- Nat was not detected on a gel but may be because a very small amount of product was made.
- Well 1,2,3 are PCR mix with Nat primers and 1.5uL of Nat ligation product per well
- Well 4 is positive control with Kan primers and Kan DNA
- Well 5 just has PCR mix with the Nat primers.
July 5
Ran PCR to amplify 100x diluted sample of URA3 positive control contained mastermix, LEU2 primers and 100x diluted LEU2 DNA. negative control contained mastermix, 100x diluted URA3
Gel Results:
Note: this PCR reaction failed, including the positive control
PCR Protocol for Plasmids Review
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 1.0 ul diluted DNA
July 7
Note: the 10x dilution of URA3 was the most effective, positive with LEU2 failed. Reran PCR reaction to amplify different concentrations of URA3. For this reaction we used a 2x, 5x, 10x, and 15x diluted samples of URA3 - ran each sample twice 2x: 1.5ul DNA, 1.5ul dH20 5x: 1ul DNA, 4ul dH20 10x: 1ul DNA, 9ul dH20 15x: 1ul DNA, 14ul dH20 positive control contained mastermix, LEU2 primers, and 20x diluted LEU2 DNA 1st negative control contained the mastermix and just LEU2 primers 2nd negative control negative contained the mastermix and just URA3 2x diluted DNA
Gel Results:
PCR Protocol for Plasmids Review
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 1.0 ul diluted DNA
Transformation:
- We are still waiting for Leu2 dropout medium to come in.
- In the meantime, we have designed an experiment to confirm that we have the pRS425 plasmid. We will cut the plasmid with specific restriction enzymes and run the digest on a gel to measure the resulting lengths.
- Of the restriction enzymes we have, we find that BamHI, XbaI, PstI, and XhoI cut the plasmid once, and EcoRI cuts the plasmid twice. We will use EcoRI and compare the lengths of the two fragments. We will also cut with BamHI and run that in a separate lane to confirm the overall plasmid length.
July 8
Transformation:
- We digested the DNA and then ran the results on a gel.
- Into a dilution microcentrifuge tube, we used 2 uL DNA (concentration 235.4 ng/uL) and 248 uL ddH20.
- BamHI tube: 2 uL diluted DNA, 2 uL NEBuffer, 2 uL diluted BSA (10x), 1 uL BamHI enzyme, 13 uL ddH20
- EcoRI tube: 2 uL diluted DNA, 2 ul SH buffer, 1 uL EcoRI enzyme, 15 uL reaction
- Incubated both tubes in a 37 C water bath for one hour for digestion
- Ran the solutions on a gel
- 5 uL loading dye into three tubes: BamHI, EcoRI, control (uncut plasmid DNA)
- Lane 1: 5 uL ladder
- Lanes 2 and 3: 5 uL each BamHI
- Lanes 4 and 5: 5 uL each EcoRI
- Lane 6: 5 uL control
- Ran it for one hour at 132 volts
July 11
PCR group: Reran five wells of PCR on URA3 - used 10x dilution URA3 DNA in our wells. Positive control contained 2ul of 20x diluted LEU2, LEU2 primers, and mastermix. Negative control contained 1ul 10x dilution URA3 and mastermix --> check for Plasmid DNA
PCR Protocol for Plasmids Review
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
- 20.5 ul/well
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 1.0 ul diluted DNA
Gel Results:
Transformation:
- Set up yeast overnight for BC178
- Ran gel again with lower concentration enzyme; it did not work again
- E. coli transformation of Leu2 and Ura3 and plated it with Leu2-deficient drop-out media we made
July 12
Transformation:
- Plates from yesterday
- Yeast transformation for BC178; we accidentally added 1 M LiAC instead of 100 uM, so we might have to redo it
- Set up E. coli overnight
July 13
PCR:
Colony PCR:
Protocol:
- Transfer a yeast colony to a solution of 0.2% SDS. The SDS page was made by adding 0.02g SDS to 10mL of water.
- Vortex for 15 sec
- Heat in PCR machine for 4 min at 90 degrees Celsius.2
- Microcentrifuge for 1 min.
- Pipet out the supernatent and store at -20 degrees Celsius.
PCR mix for 50uL reaction:
- 5uL 10x PCR buffer
- 1.5 uL 50mM MgCl2 (we used 3uL because we 25mM MgCl2)
- 1uL of 10mM dNTPs
- 2uL of 25% TritonX-100
- 0.3 uL Taq polymersase
- 31.7 uL water
- Add 43 uL of PCR mix to each PCR tube
- Then add 3 uL of both the forward and reverse primers
- Add 1uL of the DNA from yeast
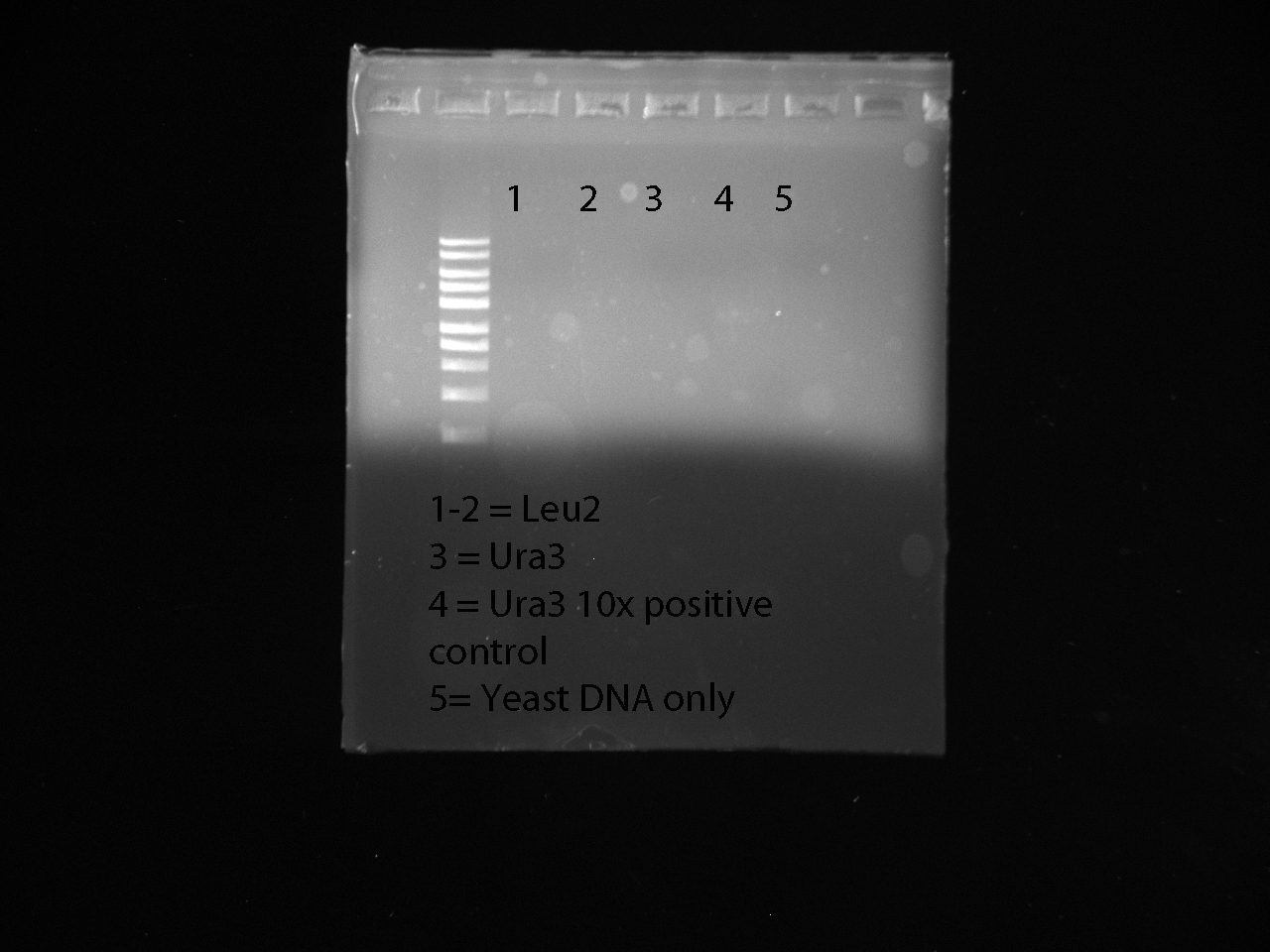
We made 2 samples with Leu2 primers, 2 samples with Uras3 primers, a positive control with Uras3 primers and DNA, and a negative control with just yeast DNA.
Strain 827 was used
Gel Results:
July 14
PCR:
Colony PCR:
Protocol:
- Transfer a yeast colony to a solution of 0.2% SDS. The SDS page was made by adding 0.02g SDS to 10mL of water.
- Vortex for 15 sec
- Heat in PCR machine for 4 min at 90 degrees Celsius.2
- Microcentrifuge for 1 min.
- Pipet out the supernatent and store at -20 degrees Celsius.
PCR mix for 50uL reaction:
- 5uL 10x PCR buffer
- 1.5 uL 50mM MgCl2 (we used 3uL because we 25mM MgCl2)
- 1uL of 10mM dNTPs
- 2uL of 25% TritonX-100. The 25% TritonX-100 was made by adding 6ul of TritonX-100 to 18ul of H20
- 0.3 uL Taq polymersase
- 31.7 uL water
- Add 43 uL of PCR mix to each PCR tube
- Then add 3 uL of both the forward and reverse primers
- Add 1uL of the DNA from yeast
We made 1 samples with Leu2 primers, 2 samples with Uras3 primers and a positive control with Uras3 primers and DNA
Strain 825 was used
Gel Results:
July 15
Ordered Biobrick primers and all 4 cassette primers without homology in order to run colony PCR
July 18
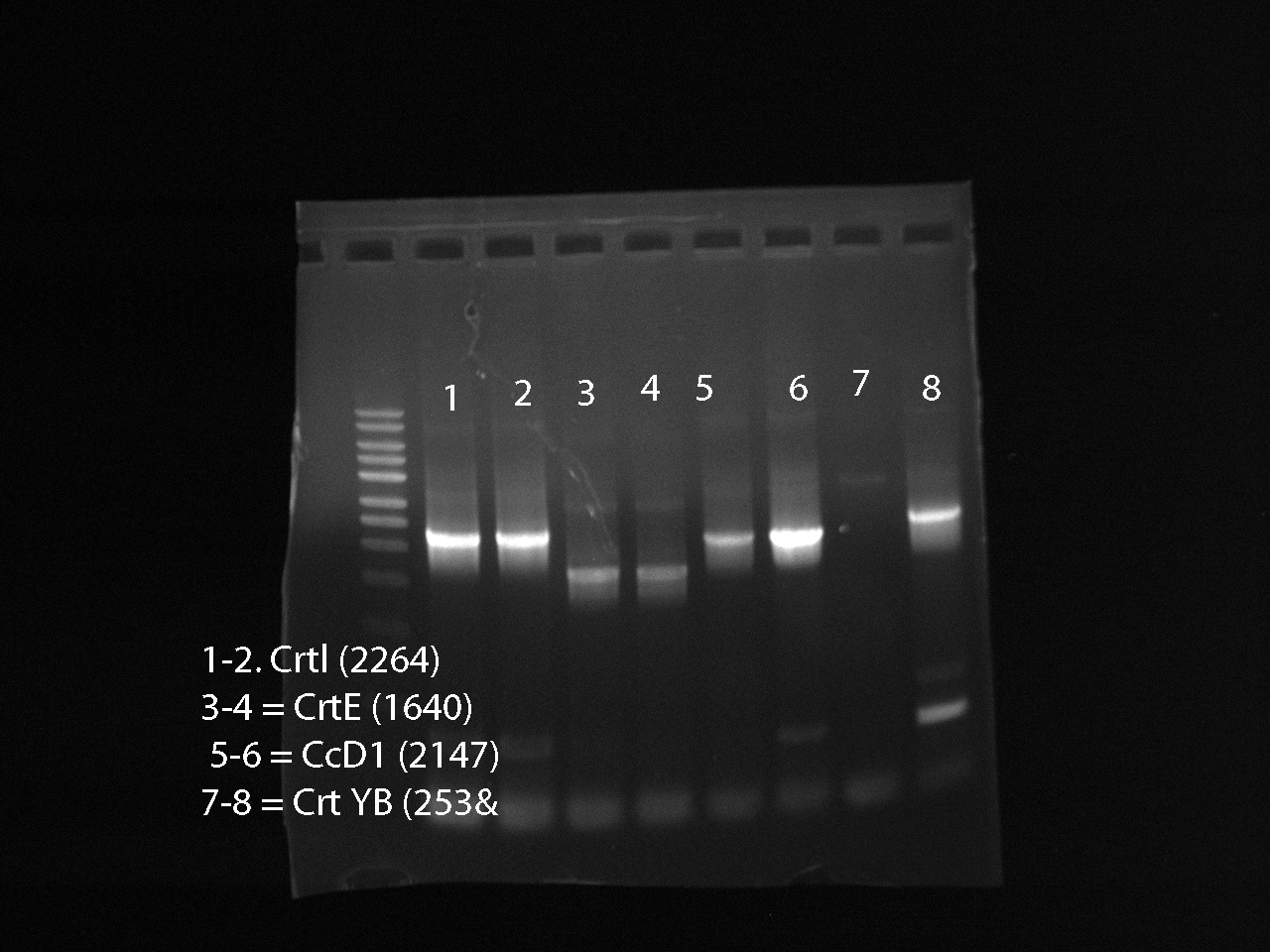
Ran a PCR for the genes: CrtI, CrtE, CcD1, CcYB --> ran 3 PCR tubes for each gene
- 1 positive control: Nat Genes + primers
- 4 negative controls, 1 corresponding to each gene, contains only the mastermix soln and F/R primers
Recipe per sample for running plasmid DNA:
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
Gel Results:
Transformation:
- We had made colonies of BC178, but we had gotten contamination in them, so we made new plates, this time with ampicillin, in hopes of getting rid of any E. coli that may have been in our test tubes. Also, we filter-sterilized our lithium acetate (just in case) and our PEG 3350.
- We set up new overnights to start over.
July 20
PCR Team: Ran a PCR for the cassettes: URA3, LEU2, KAN, NAT --> ran 2 41ul PCR tubes for each gene
- 4 25ul negative controls, 1 corresponding to each gene, contains only the mastermix soln and F/R primers
Recipe per sample for running plasmid DNA (20.5ul reactions):
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
Gel Results:
Designed primers to amplify Homology, Gene, Cassette, Homology
- crtYB-URa3
- Forward primer GAAGAATATACTAAAAAATGAGCAGGCAAGATA 33 54.78
- Reverse primer TATGAATGTCAGTAAGTATGTATACGAACAGTAT 34 54.58
- BioBrick:
- gaattcgcggccgcttctagag GAA GAA TAT ACT AAA AAA TGA GCA GGC AAG ATA
- ctgcagcggccgcractagta T ATG AAT GTC AGT AAG TAT GTA TAC GAA CAG TAT
- crtI-kanMX4
- Forward primer TTTTCCAATAGGTGGTTAGCAATCGTC 27 55.97
- Reverse primer AAATTCATAATAGAAACGACACGAAATTACAAA 33 54.14
- Biobrick:
- Gaattcgcggccgcttctagag TTT TCC AAT AGG TGG TTA GCA ATC GTC TTA
- Ctgcagcggccgcractagta AAA TTC ATA ATA GAA ACG ACA CGA AAT TAC AAA A
- CrtE-natMX4
- Forward primer TCTCCGAGCAGAAGGAAGAAC 21 53.62
- Reverse primer CTAAACTCACAAATTAGAGCTTCAATTTAATTATAT 36 52.82
- Biobricks:
- gaattcgcggccgcttctagag TCT CCG AGC AGA AGG AAG AAC
- ctgcagcggccgcractagta CTA AAC TCA CAA ATT AGA GCT TCA ATT TAA TTA TAT
- CCD1-leu2
- Forward primer AGTTACTCACTAATGACTAACGAAAAGGTCTG 32 56.97
- Reverse primer GGTTGAGCATTACGTATGATATGTCCATGT 30 57.38
- Biobricks:
- gaattcgcggccgcttctagag AGTTACTCACTAATGACTAACGAAAAGGTCTG
- ctgcagcggccgcractagta GGT TGA GCA TTA CGT ATG ATA TGT CCA TGT
Transformation:
- Made more YPD
- E. coli transformations using Nat and Kan
- Set up yeast overnights to try again. The contaminated plates must have yeast in them that could somehow grow in Leu-deficient medium. We are plating again to verify.
July 22
PCR team: Realized the reverse primer design from July 20 was wrong. The redesigned primers are as follows:
- crtYB-URa3
- Reverse primer AGTATCATACTGTTCGTATACATACTTACTGACA
- BioBrick:
- Reverse biobrick: ctgcagcggccgcractagta AGTATCATACTGTTCGTATACATACTTACTGACA
- crtI-kanMX4
- Reverse primer ATGAACATATTCCATTTTGTAATTTCGTGTC
- Biobrick:
- Reverse biobrick primer: ctgcagcggccgcractagta ATGAACATATTCCATTTTGTAATTTCGTGTC
- CrtE-natMX4
- Reverse primer GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT
- Biobrick:
- ctgcagcggccgcractagta GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT
- CCD1-leu2
- Forward primer AGTTACTCACTAATGACTAACGAAAAGGTCTG
- Reverse primer ATATTTAATTATTGTACATGGACATATCATACGTAA
- Biobricks:
- gaattcgcggccgcttctagag AGTTACTCACTAATGACTAACGAAAAGGTCTG
- ctgcagcggccgcractagta ATATTTAATTATTGTACATGGACATATCATACGTAA
 "
"