Team:ETH Zurich/Biology/Cloning
From 2011.igem.org
(Difference between revisions)
(→Design of the parts) |
|||
| Line 1: | Line 1: | ||
| - | {{:Team:ETH Zurich/Templates/Header/ | + | {{:Team:ETH Zurich/Templates/Header/Biology|currPage=Cloning}} |
Revision as of 21:15, 10 September 2011
Used parts from registry
- BBa R0040 PTet
- BBa R0051 λP
- BBa R0010 Plac
- BBa R0061 Plux
- BBa J23100 Pconst
- BBa C0061 LuxILVA
- BBa C0040 TetRLVA
- BBa C0051 CI
- BBa C0062 LuxR
- BBa B0015 Double Terminator
Synthesized parts
- LacIM1
- AlcR (codon optimized)
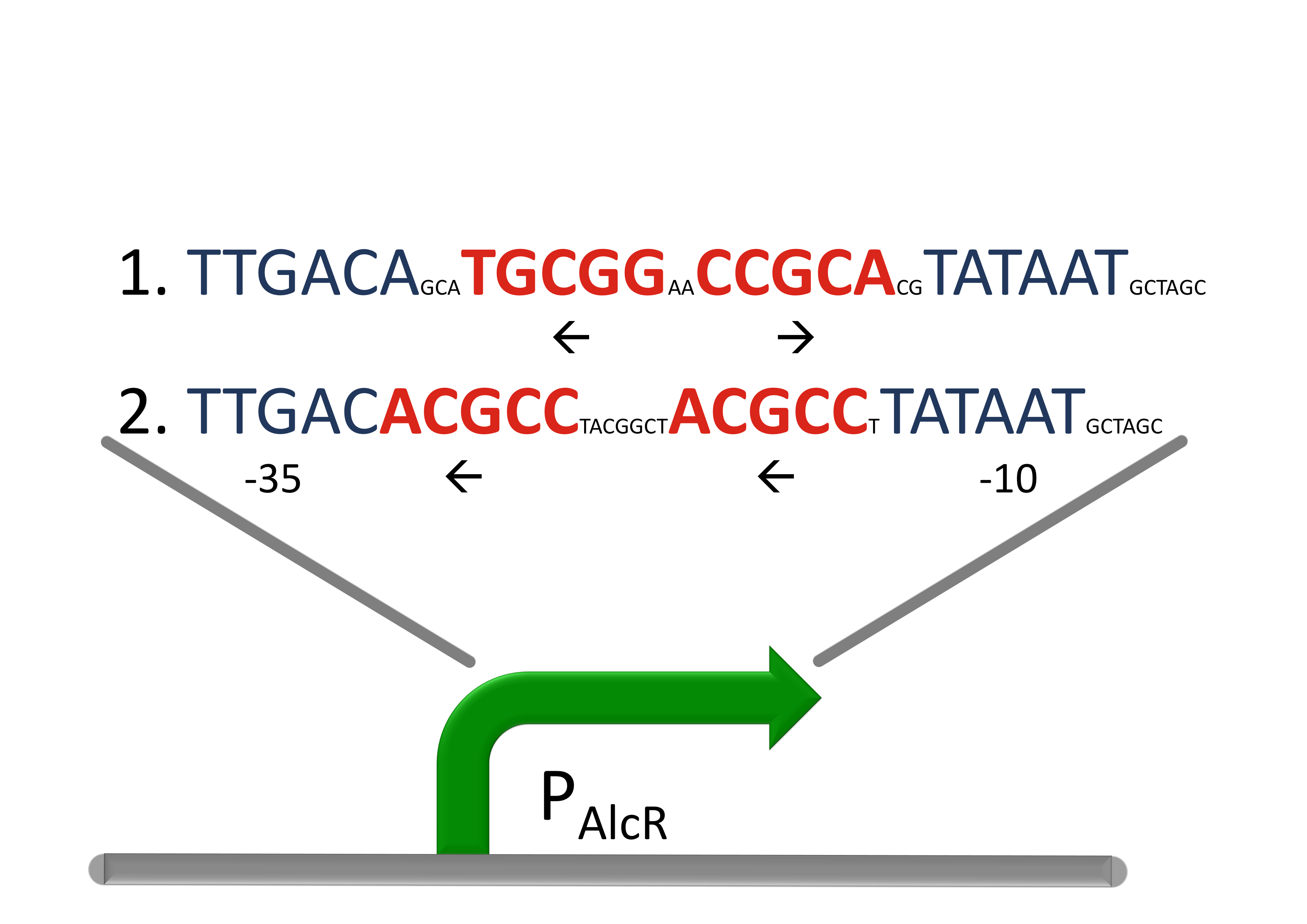
Design of a AlcR/acetaldeyhde repressed promotor PAlcR
For the promotor PAlcR the operator site of AlcR-inducer from apergillus nidulans was placed between the -10 and -35 region of the strong promotor. While acetaldeyhde is bind to AlcR, the complex will bind to the operon and block the binding of the RNA-polymerase.
Design of the test-system
To test our designed AlcR-promotors the following system was designed (see Figure). To induce the expression of AlcR a tetracycline induced promotor was used. To monitor the expression of AlcR a His-tag was introduced. In present of acetaldeyhde AlcR binds to the AlcR operon and inhibits the expression of GFP. For a better signal assam gfp was used.
 "
"