Team:Paris Liliane Bettencourt/Notebook/2011/08/11/
From 2011.igem.org

Contents |
Cyrille
After a long analysis, we discovered that we were working with the wrong plasmid map. Then the interpretation of the manipulations become more clear. Here is the new plasmid map:
Then, we reanalyze the results of the manipulation of the 05/08 and discovered that we may had the good results.
The tubes of 5 and 6, that where identified to be the uncorrect sequence this time are supposed to be the good ones. They where recollected from the trash bin.
To check the presence of the expected plasmid in these tube we will proceed to a control digestion. This time, we want to verify the presence of all the interesting restriction sites. Then we will load a gel.
0.5 µg of DNA from the tubes will be digested into:
a - No digestion
-> one band expected at 5.9kb
-> verify the presence of the plasmid and the pollution of the MP
b - EcoRI (NEB)
-> one band expected at 5.9kb (a bit upper than the previous one, because it is linear)
-> Verify the presence only one EcoRI site in the plasmid
c - EcoRI and EcoRV (NEB)
-> One band at 839bp and one at 5,1kb (if uncorrect muation 2.2kb and 3.7kb) (if other map with "correct": 2.2kb and 3.7kb and "uncorrect": 5.08kb 839bp)
-> Verify that the good EcoRI has been eliminated
d - PstI and EcoRV (NEB)
-> One band at 5,05kb and one at 878bp (idem for the uncorrect mutation) (if other map with both "correct" and "uncorrect" mutation 2.3kb and 3.7kb)
-> Verify that the plasmid is assembled the way we think it is
-> Verify that the PstI site is functionnal
PstI, EcoRI and PstI works in NEBuffer3. The digestion will be done for one hour at 37°C in 10µL
The tube composition is: Tube 6: 203 ng/µL (ratio 2,1) Tube 7: 480 ng/µL (ratio 2,22)
Due to the bad ratio of the second tube, there are probably proteins, and the quantity of DNA is probably underestimated. We will consider it as the fisrt tube.
Then:
| a | b | c | d | |
| DNA | 2,5µL | 2,5µL | 2,5µL | 2,5µL |
| NEBuffer3 | . | 1µL | 1µL | 1µL |
| H20 | 7,5µL | 5,5µL | 4,5µL | 4,5µL |
| EcoRI | . | 1µL | 1µL | . |
| EcoRV | . | . | 1µL | 1µL |
| PstI | . | . | . | 1µL |
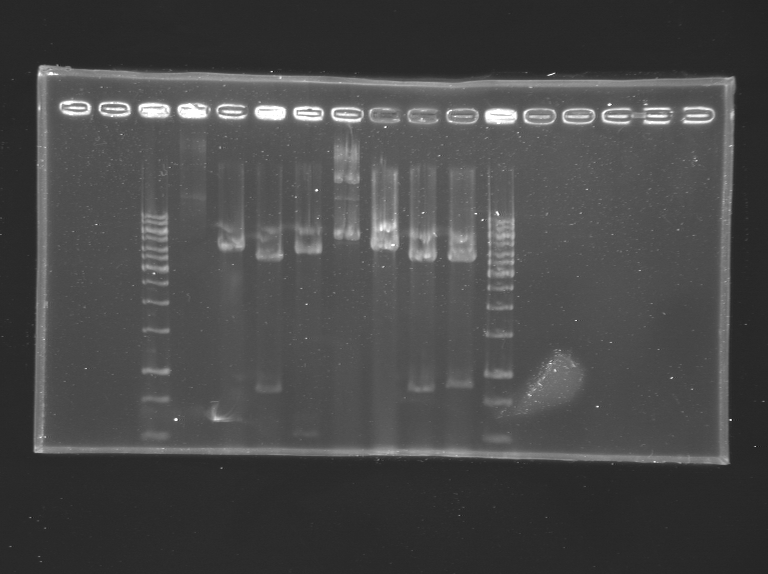
The digestion has been held 2 hours. Then the results has been loaded on a gel. The gel is not clean but the result for the tube 6 is the result expected.
The band of the line 6 has been extracted and will be purified.
The Gel extraction and the original miniprep sample has been transformed and plated. The results were analyszed the day after.
Kevin
TECAN from PCR Purification DL Plasmids
- 1=LacI:GFP
1.4 : 176,7 ng/uL - 1.5 : 179,2 ng/uL
- 2=tetR:YFP
2.4 : 181,5 ng/uL - 1.5 : 138,4 ng/uL
- 3=LacO array
3.7 : 109,8 ng/uL - 3.8 : 105,8 ng/uL
- 4=tetO array
4.7 : 85 ng/uL - 4.8 : 92 ng/uL
Digestion
Digestion of plasmids with Fast Digest XS restriction enzyme.
0,2ng PCR Product
 "
"