Team:Paris Liliane Bettencourt/Notebook/2011/08/04/
From 2011.igem.org

Contents |
Cyrille
Elimitation of the EcoRI site by the EcoRI-Klenow method
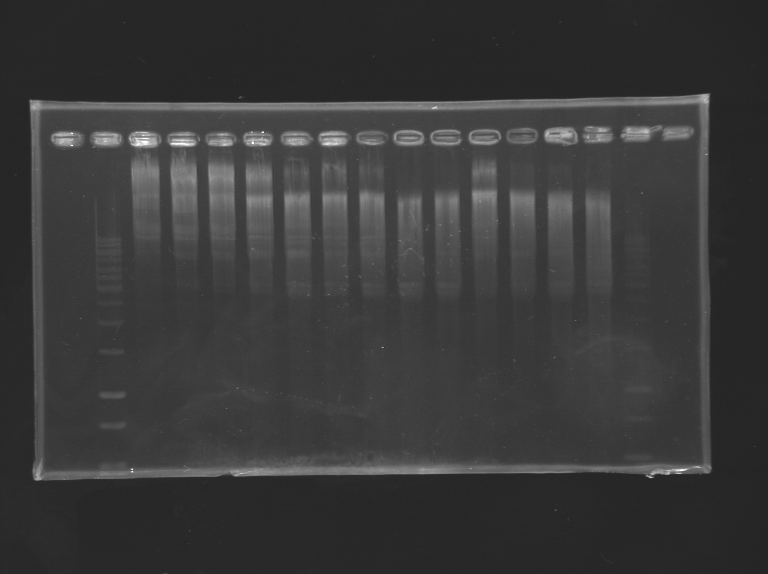
The template will overgo a menaged digestion with normal EcoRI. 12 tubes of 2 µg of DNA template will be prepared, in the fastdigest buffer. They will be digested with EcoRI for different duration: 0 - 1 - 3 - 5 - 6 - 7 - 8 - 9 - 10 - 11 - 13 - 15 min
The inactivation of the enzyme is done by eating 5 min at 80°C.
The results are loaded on a gel.
The bands 3-5 are cutted together, as well as 6,7,8,9, and 10-11. They will undergo a gel extraction.
The final concentration is 21.2, 23.5 and 30.8 ng/µL
Danyel & Camille
T7 amber
According to the T7 amber sequencing results the S15 strain only held the terminator and not the T7 polymerase. The cloning step of the T7 + standard terminator has to be redone. An overnight quickmut PCR was launched: 4 tubes in total: negative control without dNTP and 120 ng of template, tube with 20ng of template, 40ng, and 120 ng.
tRNA
Transformation of tRNA +terminator.
Kevin
Check of PCR with gel
- 3,5uL Loading Dye 6X
- 16,5uL PCR Mix or 1uL MP of Dave Lane Plasmid + 15uL of water DNAase free
TECAN of S27
I forget to write which of my eppendorfs are my witness. Tecan aren't enough to know, so I have to do a gel.
 "
"