Vitamin C
Models
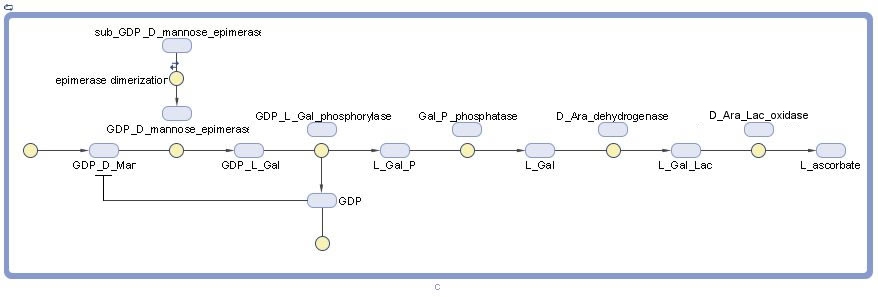
Graphical model
LBS model
Our full LBS model includes both gene expression and the metabolic pathway.
The full pathway+expression l-ascorbate model in LBS
We developed a simplified LBS model to examine the metabolic pathway in isolation from gene expression.
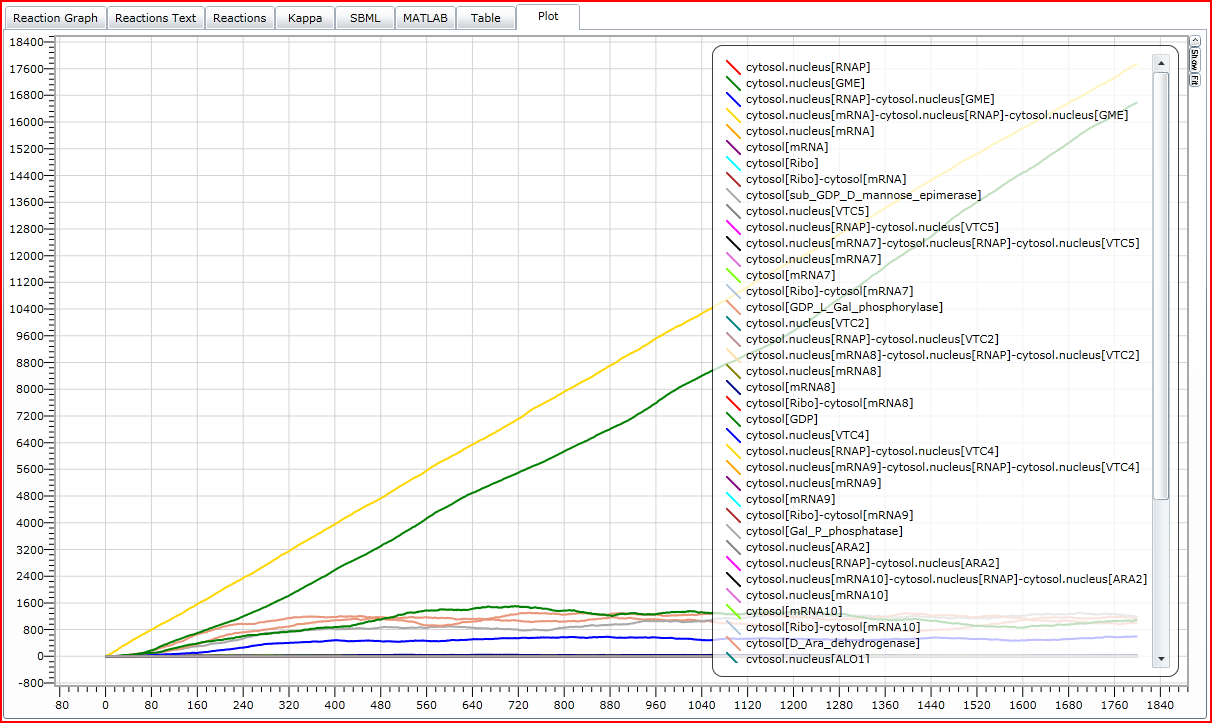
Matlab model
We chose to port our model to Matlab's SimBiology Toolbox in order to facilitate complex analysis. To speed computations, this model has been simplified by taking enzyme concentrations as constant, assuming that the gene expression component of our system has reached a steady state.
The full l-ascorbate model as exported from the Matlab SimBiology Toolbox
Note: all parameters for this model listed here.
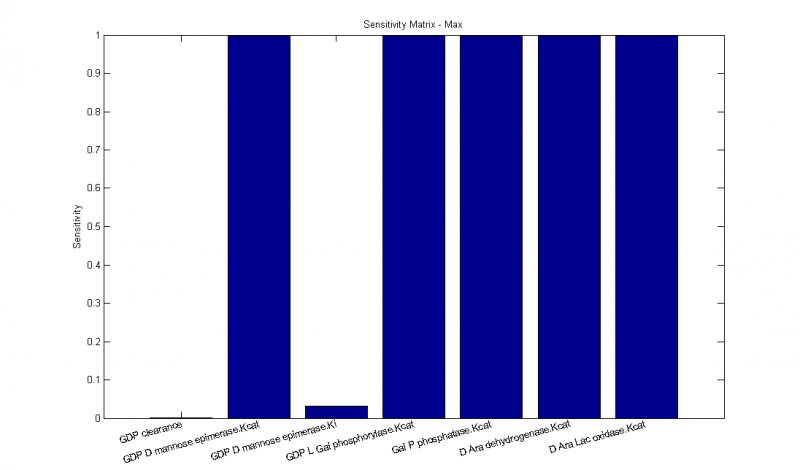
Sensitivity Analysis
The model is equally sensitive to small changes in all kcat values (the maximum turnover rate of saturated enzyme). This is reasonable, since our pathway is fairly simplistic and linear. A decrease in any enzyme's turnover rate will cause a bottleneck. We notice, however, that the model is significantly less sensitive to the mannose epimerase inhibition binding constant, ki. Even though ki is an inhibition constant, the corresponding sensitivity \[d[l-ascorbate]/d[k_{i}]\] is positive, since increasing ki means that more inhibitor is requires to yield the same degree of inhibition, much like how increasing km means that more substrate is needed to drive the equilibrium towards the enzyme-substrate complex. The GDP clearance rate also has little effect on the model, suggesting that the pathway is producing a low enough amount of GDP that it can be cleared before it can inhibit the epimerase. Alternatively, it might just suggest that whatever the amount of GDP present, the effect on mannose epimerase is minor. In either case, the parameter sensitivities suggest that inhibition is of little importance in this model, at least at the substrate influx and enzyme production levels we are working with.Optimization
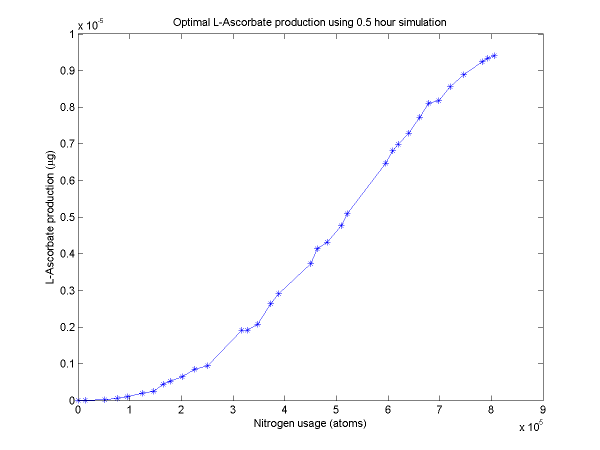
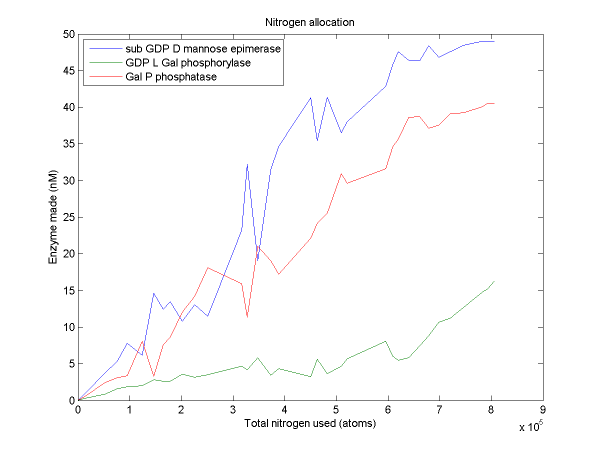
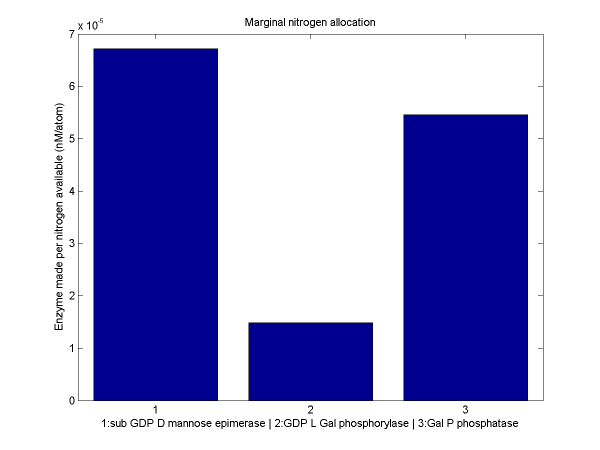
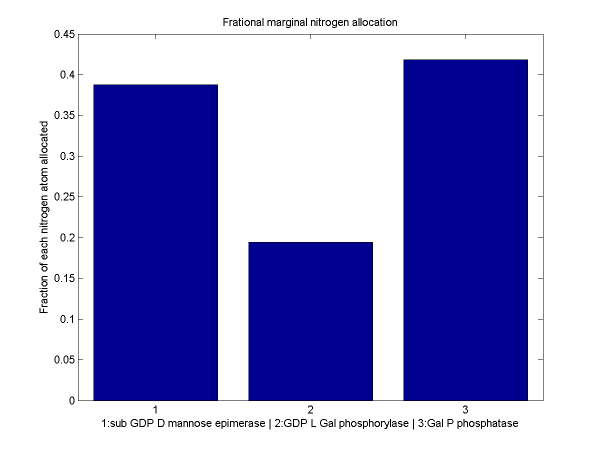
For a discussion of the optimization techniques used and the nitrogen allocation analysis which follows, see our optimization techniques.
 "
"