Team:Johns Hopkins/Data
From 2011.igem.org
Data Page
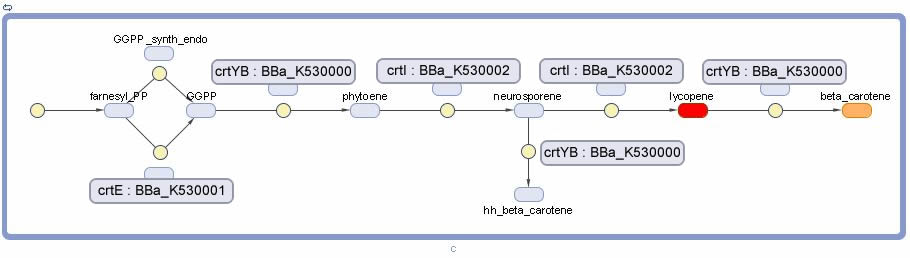
Vitamin A Production
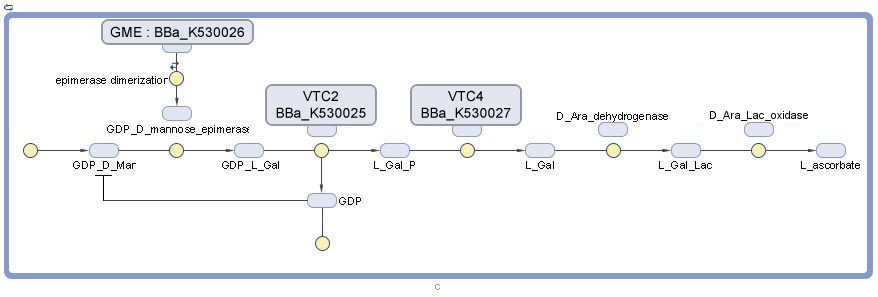
Vitamin C Production
Yeast Toolkit
|
Integrating vectors |
Favorite New Parts
Vitamin A Production
- CrtYB Enzyme in the pathway required for B-Carotene Synthesis. This enzyme is a fusion, it is the full Phytoene Synthase enzyme spliced with a Lycopene B-Cyclase enzymatic domain. This sequence was taken from a WT strain of xanthophyllomyces dendrorhous. It catalyzes both the conversion of Geranylgeranyl diphosphate to Phytoene and Lycopene to B-Carotene.
- CrtE Enzyme in the pathway required for B-Carotene Synthesis. This enzyme is named Geranylgeranyl Diphosphate Synthase. This sequence was taken from a WT strain of xanthophyllomyces dendrorhous. It catalyzes both the conversion of Geranylgeranyl diphosphate to Phytoene and Lycopene to B-Carotene.
- CrtI Enzyme in the pathway required for B-Carotene Synthesis. This enzyme is named Phytoene Desaturase. This sequence was taken from a WT strain of xanthophyllomyces dendrorhous. It catalyzes the conversion of Phytoene to Lycopene.
Vitamin C Production
- VTC2 Enzyme in the pathway required for L-Ascorbate Acid Synthesis. This sequence was taken from plants. It catalyzes the conversion of GDP-L-Galactose to L-Galactose-1-P.
- GME Enzyme in the pathway required for L-Ascorbate Acid Synthesis. This sequence was taken from plants. It catalyzes the conversion of L-Galactose-1-P to L-Galactose.
- VTC4 Enzyme in the pathway required for L-Ascorbate Acid Synthesis. This sequence was taken from plants. It catalyzes the conversion of L-Galactose to L-Galactono-1,4-Lactone.
Promoters
- KRE9p Promoter used to regulate the expression of genes within the genome of Saccharomyces Cerevisiae or baker's yeast.
- TDH3p Promoter used to regulate the expression of genes within the genome of Saccharomyces Cerevisiae or baker's yeast.
Terminators
- HHO1utr Terminator is used to regulate the expression of genes within the genome of Saccharomyces Cerevisiae or baker's yeast.
- ARD1utr Terminator is used to regulate the expression of genes within the genome of Saccharomyces Cerevisiae or baker's yeast.
Vectors
- pRSBB413 HIS3 CEN/ARS Yeast Shuttle Vector Used for the propagation in E. coli and transformation into S. cerevisiae. Contains an Ampicillin resistance marker for E. coli and HIS3 selectable marker for S. cerevisiae. Fully compatible for use with Standard Assembly. Contains EcoRI, XbaI, SpeI, and PstI in the multiple cloning site, in that order. This vector is a CEN/ARS episomal yeast vector.
- pRSBB426 URA3 2Micron Episomal Yeast Shuttle Vector Used for propagation in E. coli and transformation into S. cerevisiae. Contains an Ampicillin resistance marker for E. coli and URA3 selectable marker for S. cerevisiae. Contains EcoRI, NheI, SpeI, and PstI in the multiple cloning site, in that order. For assembly, this vector differs from a Standard Assembly vector by replacing XbaI with NheI. This vector is a 2micron episomal yeast vector.
Existing Parts Characterized
- Gal1p. This characterization is a work in progress. We will update the part's experience page after the regional jamboree. We are working with the British Columbia team on troubleshooting.
All Submitted Parts
 "
"




