Team:Groningen/Project
From 2011.igem.org
Project description - circuit design and functionality
The aim of our project is to create a genetic device working in E.coli that will be able to count and memorize the occurrences of an input signal. This functionality will be achieved by the utilization of autoinducing loops, that will act as memory units, and interfering taRNA-crRNA system, which will act as an AND gate. Each increase of the counter will result in a different state of the system, that will result in an output signal.
The design of the device is modular. This means that both the input signal and the output signals can be changed freely, without disturbing the functionality of the memory system. Also, the design allows implementation of any number of memory units, as the AND gate design allows for a nearly hassle-free extension of the system, provided enough functional memory units.
Because of that, the range of applications of our system can vary from the bacteria being used as a memory system of a Turing complete machine, through bacteria used as biosensors measuring the number of occurrences of an event in a process, to bacteria that can perform multi-step bioconversions sequentially all by themselves and without the need of changing them to another strain.
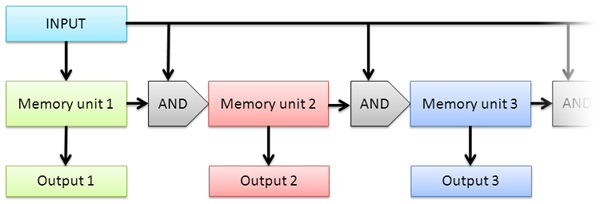
Figure 1 - The outline of the project. The first occurrence of the input signal triggers the first memory unit (which consists of an autoinducing loop and a jammer-based off switch) to start. The first memory unit starts triggering the first AND gate. The second occurrence of the input triggers the AND gate. If the first memory unit is running, the AND gate triggers the second memory unit (which also consists of an autoinducing loop and a jammer-based off switch) to start. The second module starts triggering the second AND gate and represses the first module with the use of the jammer-based off switch. This process can be continued, if more autoinducing loops are provided. During operation, the system produces an output which depends on the current running memory unit.
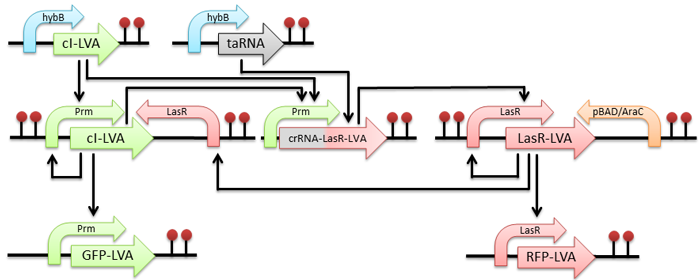 Figure 2 - The design of the circuit. We have chosen to use the hybB operon promoter as our input signal. This promoter reacts, among other signals, to a cold shock.
Figure 2 - The design of the circuit. We have chosen to use the hybB operon promoter as our input signal. This promoter reacts, among other signals, to a cold shock.
The first occurrence of the input signal causes the cell to produce cI-LVA protein (which activates the modified Prm promoter) and taRNA (which has no effect before it degrades). The cI-LVA activates three operons: first, which is an autoinducing loop creating more cI-LVA protein, second which produces signal - GFP, and third which produces the crRNA-LasR-LVA transcript. This transcript is self-inhibiting itself.
The second occurence of the input signal produces taRNA again, which now can bind to the crRNA part of the crRNA-LasR-LVA transcript. It activates the transcript and leads to creation of LasR-LVA protein. This protein activates two operons: first which is an autoinducing loop creating more LasR-LVA protein and second which produces signal - RFP. The third effect of LasR-LVA protein is shutting down of the first autoinducing loop via a jammer - LasR promoter placed on the anti-sense strand of DNA after the operon.
The system has been divided into three fully functional sub-modules: the cI-based autoinducing loop, the LasR-based autoinducing loop and the taRNA-caRNA-based AND gate. Different variants of those sub-modules will be made and tested. Upon completion, they will be combined to create a fully functional system.
Project description - modeling
Evolutionary model
Exact values of parameters for a model are hard to come by. While curated databases of models exist, the most important parameters are often specific to a particular model, environment or even differ per chassis. It is for these reasons why we have decided to extract our parameters from experimental data by the means of a evolutionary algorithm.
Our system uses parameters setting, a model description and its associated experimental data to generate a score. This score contains information about how good the model fits the experimental data with the use of those parameters. Scores from all used models and all experiments for the same parameters setting are collected and weighted. Finally, they are combined into a score for a specific parameters setting. In this way we can generate a fitness score for every parameters setting. These fitness scores can be used to drive an evolutionary model, which will find the parameters that provide the best fit for all the experimental data and for every model. As this process is quite expensive computationally, we have decided to use and outside cloud computing system. This cloud based system will be freely accessible to all iGEM teams.
Bistability model
The parameters that we will acquire in our evolutionary model will be used in our bistability model. Bistable autoinducing loops are tricky to design and build.
The key for success is that the degradation rate of the inducing factor needs to equal its production at three separate levels. Although promoters governed by Hill-kinetics allow the bistability to happen, the combination of the degradation rate of the inducer and promoter type needs to be exactly right. The modeling will help us to predict the proper solutions and change our system accordingly.
For our system, the stability of our information stored in the memory units over time is very important. In order to look more deeply into this, we will design a Markov model that will inform us about the time it takes for the information in the bacteria to degrade.
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
 "
"